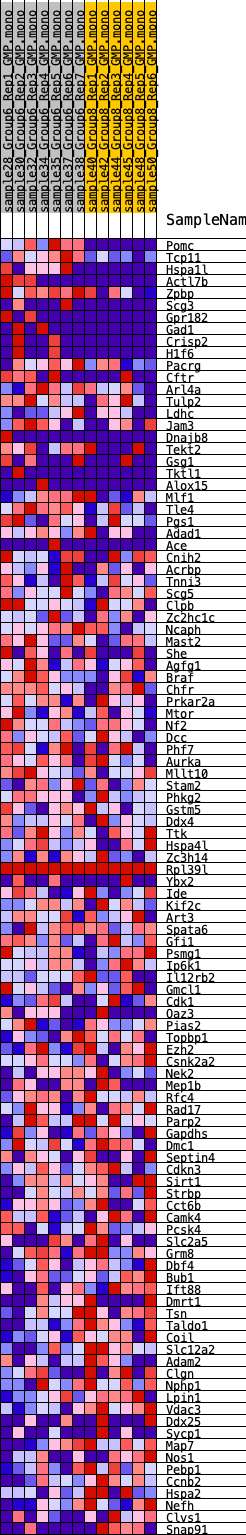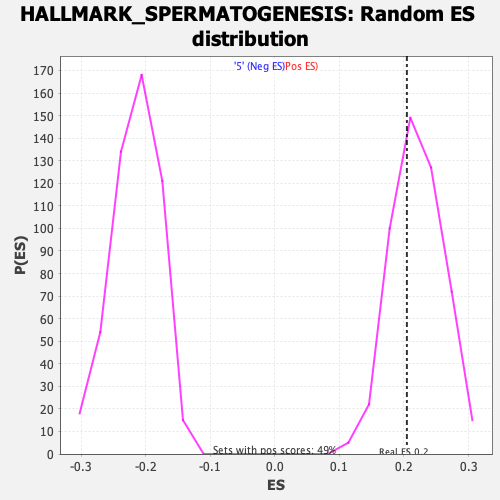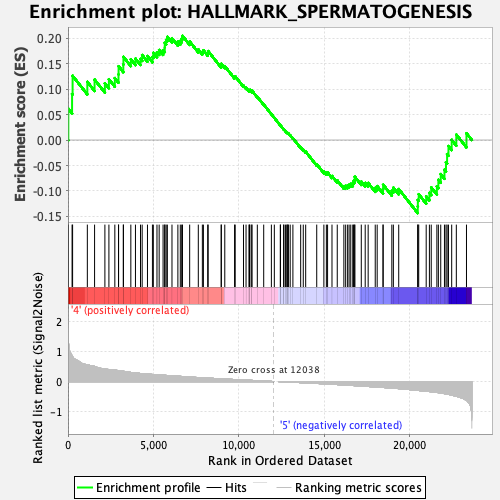
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group6_versus_Group8.GMP.mono_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | GMP.mono_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | 0.2044888 |
| Normalized Enrichment Score (NES) | 0.93089956 |
| Nominal p-value | 0.6591837 |
| FDR q-value | 0.614462 |
| FWER p-Value | 0.999 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pomc | 19 | 1.391 | 0.0616 | Yes |
| 2 | Tcp11 | 239 | 0.847 | 0.0904 | Yes |
| 3 | Hspa1l | 269 | 0.823 | 0.1261 | Yes |
| 4 | Actl7b | 1131 | 0.551 | 0.1142 | Yes |
| 5 | Zpbp | 1555 | 0.499 | 0.1186 | Yes |
| 6 | Scg3 | 2158 | 0.409 | 0.1114 | Yes |
| 7 | Gpr182 | 2393 | 0.399 | 0.1193 | Yes |
| 8 | Gad1 | 2740 | 0.379 | 0.1216 | Yes |
| 9 | Crisp2 | 2954 | 0.362 | 0.1288 | Yes |
| 10 | H1f6 | 2960 | 0.362 | 0.1448 | Yes |
| 11 | Pacrg | 3235 | 0.340 | 0.1484 | Yes |
| 12 | Cftr | 3244 | 0.339 | 0.1633 | Yes |
| 13 | Arl4a | 3676 | 0.301 | 0.1585 | Yes |
| 14 | Tulp2 | 3946 | 0.283 | 0.1598 | Yes |
| 15 | Ldhc | 4241 | 0.266 | 0.1592 | Yes |
| 16 | Jam3 | 4339 | 0.262 | 0.1669 | Yes |
| 17 | Dnajb8 | 4649 | 0.249 | 0.1649 | Yes |
| 18 | Tekt2 | 4939 | 0.234 | 0.1631 | Yes |
| 19 | Gsg1 | 4992 | 0.231 | 0.1713 | Yes |
| 20 | Tktl1 | 5205 | 0.223 | 0.1723 | Yes |
| 21 | Alox15 | 5333 | 0.220 | 0.1768 | Yes |
| 22 | Mlf1 | 5562 | 0.212 | 0.1766 | Yes |
| 23 | Tle4 | 5661 | 0.207 | 0.1817 | Yes |
| 24 | Pgs1 | 5663 | 0.207 | 0.1910 | Yes |
| 25 | Adad1 | 5744 | 0.202 | 0.1966 | Yes |
| 26 | Ace | 5813 | 0.199 | 0.2027 | Yes |
| 27 | Cnih2 | 6083 | 0.192 | 0.1999 | Yes |
| 28 | Acrbp | 6428 | 0.177 | 0.1932 | Yes |
| 29 | Tnni3 | 6570 | 0.171 | 0.1949 | Yes |
| 30 | Scg5 | 6652 | 0.168 | 0.1989 | Yes |
| 31 | Clpb | 6698 | 0.166 | 0.2045 | Yes |
| 32 | Zc2hc1c | 7118 | 0.150 | 0.1934 | No |
| 33 | Ncaph | 7623 | 0.131 | 0.1778 | No |
| 34 | Mast2 | 7860 | 0.123 | 0.1733 | No |
| 35 | She | 7921 | 0.121 | 0.1762 | No |
| 36 | Agfg1 | 8175 | 0.113 | 0.1705 | No |
| 37 | Braf | 8198 | 0.112 | 0.1746 | No |
| 38 | Chfr | 8962 | 0.089 | 0.1461 | No |
| 39 | Prkar2a | 8969 | 0.088 | 0.1498 | No |
| 40 | Mtor | 9172 | 0.083 | 0.1450 | No |
| 41 | Nf2 | 9744 | 0.066 | 0.1237 | No |
| 42 | Dcc | 9781 | 0.065 | 0.1250 | No |
| 43 | Phf7 | 10267 | 0.051 | 0.1067 | No |
| 44 | Aurka | 10416 | 0.046 | 0.1025 | No |
| 45 | Mllt10 | 10587 | 0.042 | 0.0971 | No |
| 46 | Stam2 | 10618 | 0.041 | 0.0977 | No |
| 47 | Phkg2 | 10642 | 0.040 | 0.0985 | No |
| 48 | Gstm5 | 10754 | 0.037 | 0.0955 | No |
| 49 | Ddx4 | 10757 | 0.037 | 0.0971 | No |
| 50 | Ttk | 11076 | 0.028 | 0.0848 | No |
| 51 | Hspa4l | 11449 | 0.017 | 0.0697 | No |
| 52 | Zc3h14 | 11898 | 0.004 | 0.0508 | No |
| 53 | Rpl39l | 12071 | 0.000 | 0.0435 | No |
| 54 | Ybx2 | 12421 | -0.003 | 0.0288 | No |
| 55 | Ide | 12430 | -0.003 | 0.0286 | No |
| 56 | Kif2c | 12614 | -0.007 | 0.0212 | No |
| 57 | Art3 | 12631 | -0.008 | 0.0208 | No |
| 58 | Spata6 | 12729 | -0.010 | 0.0172 | No |
| 59 | Gfi1 | 12803 | -0.013 | 0.0146 | No |
| 60 | Psmg1 | 12827 | -0.013 | 0.0143 | No |
| 61 | Ip6k1 | 12864 | -0.014 | 0.0134 | No |
| 62 | Il12rb2 | 12893 | -0.015 | 0.0129 | No |
| 63 | Gmcl1 | 12999 | -0.018 | 0.0092 | No |
| 64 | Cdk1 | 13157 | -0.022 | 0.0035 | No |
| 65 | Oaz3 | 13612 | -0.035 | -0.0142 | No |
| 66 | Pias2 | 13764 | -0.040 | -0.0189 | No |
| 67 | Topbp1 | 13905 | -0.044 | -0.0228 | No |
| 68 | Ezh2 | 14551 | -0.062 | -0.0475 | No |
| 69 | Csnk2a2 | 14969 | -0.075 | -0.0618 | No |
| 70 | Nek2 | 15115 | -0.079 | -0.0645 | No |
| 71 | Mep1b | 15186 | -0.082 | -0.0638 | No |
| 72 | Rfc4 | 15444 | -0.090 | -0.0706 | No |
| 73 | Rad17 | 15752 | -0.099 | -0.0793 | No |
| 74 | Parp2 | 16139 | -0.111 | -0.0907 | No |
| 75 | Gapdhs | 16245 | -0.115 | -0.0900 | No |
| 76 | Dmc1 | 16359 | -0.119 | -0.0894 | No |
| 77 | Septin4 | 16445 | -0.122 | -0.0876 | No |
| 78 | Cdkn3 | 16542 | -0.126 | -0.0860 | No |
| 79 | Sirt1 | 16661 | -0.131 | -0.0852 | No |
| 80 | Strbp | 16700 | -0.132 | -0.0809 | No |
| 81 | Cct6b | 16771 | -0.134 | -0.0779 | No |
| 82 | Camk4 | 16779 | -0.134 | -0.0721 | No |
| 83 | Pcsk4 | 17158 | -0.148 | -0.0816 | No |
| 84 | Slc2a5 | 17387 | -0.156 | -0.0843 | No |
| 85 | Grm8 | 17560 | -0.162 | -0.0843 | No |
| 86 | Dbf4 | 17974 | -0.178 | -0.0938 | No |
| 87 | Bub1 | 18088 | -0.181 | -0.0905 | No |
| 88 | Ift88 | 18429 | -0.195 | -0.0962 | No |
| 89 | Dmrt1 | 18436 | -0.195 | -0.0877 | No |
| 90 | Tsn | 18945 | -0.216 | -0.0996 | No |
| 91 | Taldo1 | 19034 | -0.220 | -0.0935 | No |
| 92 | Coil | 19348 | -0.234 | -0.0963 | No |
| 93 | Slc12a2 | 20461 | -0.292 | -0.1305 | No |
| 94 | Adam2 | 20467 | -0.292 | -0.1176 | No |
| 95 | Clgn | 20518 | -0.295 | -0.1065 | No |
| 96 | Nphp1 | 20955 | -0.319 | -0.1107 | No |
| 97 | Lpin1 | 21143 | -0.331 | -0.1038 | No |
| 98 | Vdac3 | 21250 | -0.338 | -0.0931 | No |
| 99 | Ddx25 | 21580 | -0.360 | -0.0909 | No |
| 100 | Sycp1 | 21667 | -0.367 | -0.0781 | No |
| 101 | Map7 | 21806 | -0.379 | -0.0669 | No |
| 102 | Nos1 | 22024 | -0.400 | -0.0582 | No |
| 103 | Pebp1 | 22113 | -0.410 | -0.0435 | No |
| 104 | Ccnb2 | 22179 | -0.417 | -0.0276 | No |
| 105 | Hspa2 | 22259 | -0.426 | -0.0118 | No |
| 106 | Nefh | 22447 | -0.447 | 0.0003 | No |
| 107 | Clvs1 | 22718 | -0.486 | 0.0106 | No |
| 108 | Snap91 | 23312 | -0.620 | 0.0133 | No |