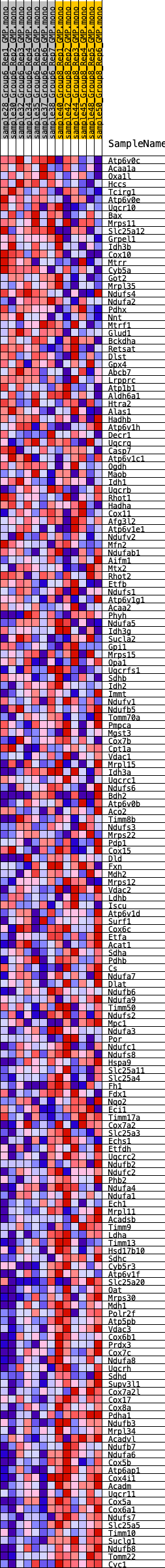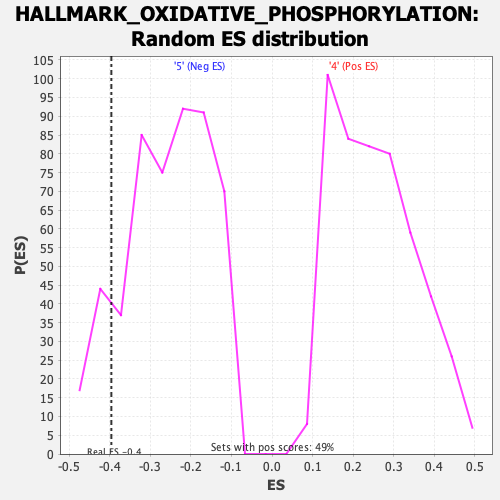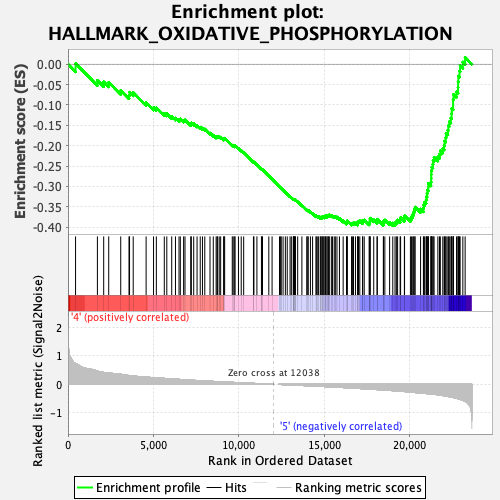
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group6_versus_Group8.GMP.mono_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | GMP.mono_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_OXIDATIVE_PHOSPHORYLATION |
| Enrichment Score (ES) | -0.39629155 |
| Normalized Enrichment Score (NES) | -1.5372506 |
| Nominal p-value | 0.12133072 |
| FDR q-value | 0.40668988 |
| FWER p-Value | 0.345 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Atp6v0c | 442 | 0.725 | 0.0018 | No |
| 2 | Acaa1a | 1720 | 0.464 | -0.0395 | No |
| 3 | Oxa1l | 2088 | 0.415 | -0.0433 | No |
| 4 | Hccs | 2382 | 0.399 | -0.0444 | No |
| 5 | Tcirg1 | 3085 | 0.354 | -0.0643 | No |
| 6 | Atp6v0e | 3569 | 0.309 | -0.0761 | No |
| 7 | Uqcr10 | 3600 | 0.307 | -0.0686 | No |
| 8 | Bax | 3815 | 0.293 | -0.0694 | No |
| 9 | Mrps11 | 4570 | 0.252 | -0.0944 | No |
| 10 | Slc25a12 | 5004 | 0.231 | -0.1063 | No |
| 11 | Grpel1 | 5168 | 0.224 | -0.1069 | No |
| 12 | Idh3b | 5633 | 0.208 | -0.1208 | No |
| 13 | Cox10 | 5780 | 0.200 | -0.1213 | No |
| 14 | Mtrr | 6068 | 0.193 | -0.1280 | No |
| 15 | Cyb5a | 6287 | 0.182 | -0.1321 | No |
| 16 | Got2 | 6495 | 0.174 | -0.1360 | No |
| 17 | Mrpl35 | 6571 | 0.171 | -0.1343 | No |
| 18 | Ndufs4 | 6764 | 0.164 | -0.1379 | No |
| 19 | Ndufa2 | 6844 | 0.161 | -0.1366 | No |
| 20 | Pdhx | 7180 | 0.147 | -0.1467 | No |
| 21 | Nnt | 7215 | 0.146 | -0.1440 | No |
| 22 | Mtrf1 | 7354 | 0.141 | -0.1459 | No |
| 23 | Glud1 | 7550 | 0.134 | -0.1504 | No |
| 24 | Bckdha | 7736 | 0.127 | -0.1547 | No |
| 25 | Retsat | 7866 | 0.123 | -0.1567 | No |
| 26 | Dlst | 8002 | 0.118 | -0.1591 | No |
| 27 | Gpx4 | 8308 | 0.109 | -0.1690 | No |
| 28 | Abcb7 | 8500 | 0.103 | -0.1743 | No |
| 29 | Lrpprc | 8666 | 0.097 | -0.1785 | No |
| 30 | Atp1b1 | 8722 | 0.095 | -0.1782 | No |
| 31 | Aldh6a1 | 8755 | 0.094 | -0.1768 | No |
| 32 | Htra2 | 8848 | 0.092 | -0.1781 | No |
| 33 | Alas1 | 8931 | 0.089 | -0.1791 | No |
| 34 | Hadhb | 9107 | 0.085 | -0.1842 | No |
| 35 | Atp6v1h | 9124 | 0.084 | -0.1824 | No |
| 36 | Decr1 | 9164 | 0.083 | -0.1817 | No |
| 37 | Uqcrq | 9610 | 0.070 | -0.1987 | No |
| 38 | Casp7 | 9665 | 0.068 | -0.1991 | No |
| 39 | Atp6v1c1 | 9761 | 0.066 | -0.2013 | No |
| 40 | Ogdh | 9763 | 0.066 | -0.1995 | No |
| 41 | Maob | 9972 | 0.059 | -0.2066 | No |
| 42 | Idh1 | 10134 | 0.055 | -0.2119 | No |
| 43 | Uqcrb | 10277 | 0.050 | -0.2166 | No |
| 44 | Rhot1 | 10850 | 0.034 | -0.2400 | No |
| 45 | Hadha | 10877 | 0.034 | -0.2401 | No |
| 46 | Cox11 | 11052 | 0.029 | -0.2467 | No |
| 47 | Afg3l2 | 11317 | 0.021 | -0.2574 | No |
| 48 | Atp6v1e1 | 11345 | 0.020 | -0.2580 | No |
| 49 | Ndufv2 | 11378 | 0.019 | -0.2588 | No |
| 50 | Mfn2 | 11748 | 0.008 | -0.2743 | No |
| 51 | Ndufab1 | 11939 | 0.002 | -0.2824 | No |
| 52 | Aifm1 | 12374 | -0.002 | -0.3008 | No |
| 53 | Mtx2 | 12439 | -0.003 | -0.3034 | No |
| 54 | Rhot2 | 12474 | -0.004 | -0.3048 | No |
| 55 | Etfb | 12582 | -0.007 | -0.3091 | No |
| 56 | Ndufs1 | 12716 | -0.010 | -0.3145 | No |
| 57 | Atp6v1g1 | 12826 | -0.013 | -0.3188 | No |
| 58 | Acaa2 | 12992 | -0.018 | -0.3253 | No |
| 59 | Phyh | 13065 | -0.020 | -0.3279 | No |
| 60 | Ndufa5 | 13177 | -0.023 | -0.3319 | No |
| 61 | Idh3g | 13194 | -0.023 | -0.3320 | No |
| 62 | Sucla2 | 13197 | -0.024 | -0.3314 | No |
| 63 | Gpi1 | 13256 | -0.025 | -0.3331 | No |
| 64 | Mrps15 | 13259 | -0.025 | -0.3325 | No |
| 65 | Opa1 | 13297 | -0.026 | -0.3333 | No |
| 66 | Uqcrfs1 | 13300 | -0.026 | -0.3326 | No |
| 67 | Sdhb | 13437 | -0.030 | -0.3376 | No |
| 68 | Idh2 | 13683 | -0.037 | -0.3470 | No |
| 69 | Immt | 13966 | -0.046 | -0.3577 | No |
| 70 | Ndufv1 | 14027 | -0.048 | -0.3589 | No |
| 71 | Ndufb5 | 14052 | -0.048 | -0.3586 | No |
| 72 | Tomm70a | 14183 | -0.052 | -0.3626 | No |
| 73 | Pmpca | 14309 | -0.055 | -0.3664 | No |
| 74 | Mgst3 | 14510 | -0.061 | -0.3732 | No |
| 75 | Cox7b | 14525 | -0.061 | -0.3721 | No |
| 76 | Cpt1a | 14611 | -0.064 | -0.3739 | No |
| 77 | Vdac1 | 14663 | -0.065 | -0.3742 | No |
| 78 | Mrpl15 | 14762 | -0.068 | -0.3764 | No |
| 79 | Idh3a | 14789 | -0.069 | -0.3756 | No |
| 80 | Uqcrc1 | 14857 | -0.071 | -0.3764 | No |
| 81 | Ndufs6 | 14863 | -0.071 | -0.3746 | No |
| 82 | Bdh2 | 14910 | -0.073 | -0.3745 | No |
| 83 | Atp6v0b | 14959 | -0.074 | -0.3744 | No |
| 84 | Aco2 | 15033 | -0.077 | -0.3754 | No |
| 85 | Timm8b | 15051 | -0.078 | -0.3739 | No |
| 86 | Ndufs3 | 15057 | -0.078 | -0.3719 | No |
| 87 | Mrps22 | 15152 | -0.080 | -0.3736 | No |
| 88 | Pdp1 | 15154 | -0.081 | -0.3713 | No |
| 89 | Cox15 | 15232 | -0.083 | -0.3723 | No |
| 90 | Dld | 15262 | -0.085 | -0.3711 | No |
| 91 | Fxn | 15285 | -0.086 | -0.3696 | No |
| 92 | Mdh2 | 15406 | -0.089 | -0.3722 | No |
| 93 | Mrps12 | 15476 | -0.091 | -0.3725 | No |
| 94 | Vdac2 | 15576 | -0.094 | -0.3741 | No |
| 95 | Ldhb | 15640 | -0.095 | -0.3740 | No |
| 96 | Iscu | 15735 | -0.098 | -0.3753 | No |
| 97 | Atp6v1d | 15880 | -0.103 | -0.3785 | No |
| 98 | Surf1 | 16097 | -0.110 | -0.3845 | No |
| 99 | Cox6c | 16293 | -0.117 | -0.3895 | No |
| 100 | Etfa | 16332 | -0.118 | -0.3878 | No |
| 101 | Acat1 | 16333 | -0.118 | -0.3844 | No |
| 102 | Sdha | 16588 | -0.128 | -0.3916 | Yes |
| 103 | Pdhb | 16650 | -0.130 | -0.3905 | Yes |
| 104 | Cs | 16702 | -0.132 | -0.3890 | Yes |
| 105 | Ndufa7 | 16817 | -0.135 | -0.3900 | Yes |
| 106 | Dlat | 16946 | -0.140 | -0.3915 | Yes |
| 107 | Ndufb6 | 16948 | -0.140 | -0.3875 | Yes |
| 108 | Ndufa9 | 16994 | -0.141 | -0.3854 | Yes |
| 109 | Timm50 | 17071 | -0.144 | -0.3846 | Yes |
| 110 | Ndufs2 | 17235 | -0.151 | -0.3872 | Yes |
| 111 | Mpc1 | 17239 | -0.151 | -0.3831 | Yes |
| 112 | Ndufa3 | 17330 | -0.154 | -0.3825 | Yes |
| 113 | Por | 17617 | -0.165 | -0.3900 | Yes |
| 114 | Ndufc1 | 17658 | -0.166 | -0.3870 | Yes |
| 115 | Ndufs8 | 17665 | -0.166 | -0.3825 | Yes |
| 116 | Hspa9 | 17671 | -0.167 | -0.3780 | Yes |
| 117 | Slc25a11 | 17886 | -0.175 | -0.3821 | Yes |
| 118 | Slc25a4 | 18081 | -0.181 | -0.3853 | Yes |
| 119 | Fh1 | 18094 | -0.182 | -0.3806 | Yes |
| 120 | Fdx1 | 18449 | -0.196 | -0.3901 | Yes |
| 121 | Nqo2 | 18457 | -0.196 | -0.3848 | Yes |
| 122 | Eci1 | 18536 | -0.199 | -0.3825 | Yes |
| 123 | Timm17a | 18815 | -0.210 | -0.3884 | Yes |
| 124 | Cox7a2 | 19001 | -0.219 | -0.3901 | Yes |
| 125 | Slc25a3 | 19132 | -0.223 | -0.3893 | Yes |
| 126 | Echs1 | 19211 | -0.227 | -0.3861 | Yes |
| 127 | Etfdh | 19282 | -0.230 | -0.3826 | Yes |
| 128 | Uqcrc2 | 19431 | -0.237 | -0.3821 | Yes |
| 129 | Ndufb2 | 19459 | -0.239 | -0.3765 | Yes |
| 130 | Ndufc2 | 19679 | -0.250 | -0.3787 | Yes |
| 131 | Phb2 | 19689 | -0.250 | -0.3720 | Yes |
| 132 | Ndufa4 | 20032 | -0.269 | -0.3789 | Yes |
| 133 | Ndufa1 | 20099 | -0.273 | -0.3739 | Yes |
| 134 | Ech1 | 20169 | -0.277 | -0.3690 | Yes |
| 135 | Mrpl11 | 20223 | -0.280 | -0.3633 | Yes |
| 136 | Acadsb | 20257 | -0.282 | -0.3567 | Yes |
| 137 | Timm9 | 20311 | -0.286 | -0.3508 | Yes |
| 138 | Ldha | 20626 | -0.302 | -0.3556 | Yes |
| 139 | Timm13 | 20792 | -0.312 | -0.3537 | Yes |
| 140 | Hsd17b10 | 20811 | -0.314 | -0.3456 | Yes |
| 141 | Sdhc | 20863 | -0.317 | -0.3387 | Yes |
| 142 | Cyb5r3 | 20951 | -0.319 | -0.3333 | Yes |
| 143 | Atp6v1f | 20968 | -0.320 | -0.3249 | Yes |
| 144 | Slc25a20 | 21005 | -0.323 | -0.3173 | Yes |
| 145 | Oat | 21023 | -0.324 | -0.3088 | Yes |
| 146 | Mrps30 | 21080 | -0.328 | -0.3018 | Yes |
| 147 | Mdh1 | 21082 | -0.328 | -0.2925 | Yes |
| 148 | Polr2f | 21234 | -0.337 | -0.2894 | Yes |
| 149 | Atp5pb | 21247 | -0.338 | -0.2803 | Yes |
| 150 | Vdac3 | 21250 | -0.338 | -0.2707 | Yes |
| 151 | Cox6b1 | 21255 | -0.339 | -0.2613 | Yes |
| 152 | Prdx3 | 21286 | -0.341 | -0.2528 | Yes |
| 153 | Cox7c | 21346 | -0.346 | -0.2455 | Yes |
| 154 | Ndufa8 | 21360 | -0.347 | -0.2362 | Yes |
| 155 | Uqcrh | 21417 | -0.351 | -0.2286 | Yes |
| 156 | Sdhd | 21638 | -0.365 | -0.2276 | Yes |
| 157 | Supv3l1 | 21734 | -0.372 | -0.2210 | Yes |
| 158 | Cox7a2l | 21782 | -0.377 | -0.2123 | Yes |
| 159 | Cox17 | 21927 | -0.391 | -0.2073 | Yes |
| 160 | Cox8a | 22006 | -0.398 | -0.1993 | Yes |
| 161 | Pdha1 | 22027 | -0.400 | -0.1888 | Yes |
| 162 | Ndufb3 | 22098 | -0.407 | -0.1802 | Yes |
| 163 | Mrpl34 | 22119 | -0.410 | -0.1694 | Yes |
| 164 | Acadvl | 22224 | -0.422 | -0.1618 | Yes |
| 165 | Ndufb7 | 22253 | -0.425 | -0.1509 | Yes |
| 166 | Ndufa6 | 22308 | -0.430 | -0.1410 | Yes |
| 167 | Cox5b | 22398 | -0.440 | -0.1323 | Yes |
| 168 | Atp6ap1 | 22440 | -0.446 | -0.1213 | Yes |
| 169 | Cox4i1 | 22449 | -0.448 | -0.1089 | Yes |
| 170 | Acadm | 22529 | -0.459 | -0.0992 | Yes |
| 171 | Uqcr11 | 22530 | -0.459 | -0.0862 | Yes |
| 172 | Cox5a | 22554 | -0.463 | -0.0740 | Yes |
| 173 | Cox6a1 | 22730 | -0.487 | -0.0676 | Yes |
| 174 | Ndufs7 | 22819 | -0.501 | -0.0571 | Yes |
| 175 | Slc25a5 | 22824 | -0.501 | -0.0430 | Yes |
| 176 | Timm10 | 22839 | -0.504 | -0.0292 | Yes |
| 177 | Suclg1 | 22904 | -0.518 | -0.0172 | Yes |
| 178 | Ndufb8 | 22940 | -0.525 | -0.0037 | Yes |
| 179 | Tomm22 | 23099 | -0.555 | 0.0053 | Yes |
| 180 | Cyc1 | 23233 | -0.598 | 0.0167 | Yes |