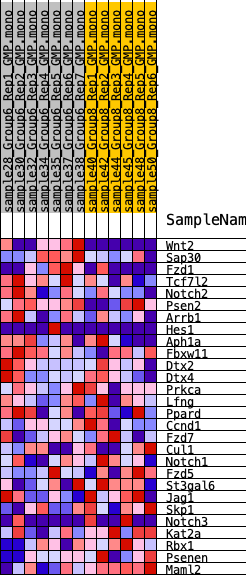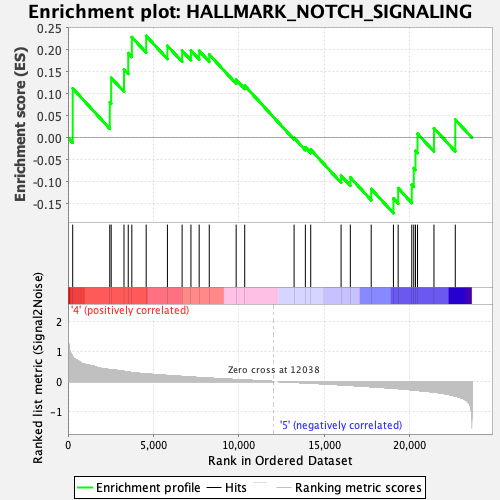
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group6_versus_Group8.GMP.mono_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | GMP.mono_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_NOTCH_SIGNALING |
| Enrichment Score (ES) | 0.23113854 |
| Normalized Enrichment Score (NES) | 0.72275364 |
| Nominal p-value | 0.8133595 |
| FDR q-value | 0.92845565 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Wnt2 | 275 | 0.816 | 0.1121 | Yes |
| 2 | Sap30 | 2444 | 0.397 | 0.0803 | Yes |
| 3 | Fzd1 | 2527 | 0.389 | 0.1359 | Yes |
| 4 | Tcf7l2 | 3276 | 0.336 | 0.1552 | Yes |
| 5 | Notch2 | 3524 | 0.312 | 0.1921 | Yes |
| 6 | Psen2 | 3734 | 0.298 | 0.2284 | Yes |
| 7 | Arrb1 | 4573 | 0.252 | 0.2311 | Yes |
| 8 | Hes1 | 5816 | 0.199 | 0.2087 | No |
| 9 | Aph1a | 6676 | 0.167 | 0.1976 | No |
| 10 | Fbxw11 | 7192 | 0.147 | 0.1980 | No |
| 11 | Dtx2 | 7674 | 0.129 | 0.1972 | No |
| 12 | Dtx4 | 8264 | 0.110 | 0.1889 | No |
| 13 | Prkca | 9838 | 0.063 | 0.1318 | No |
| 14 | Lfng | 10338 | 0.048 | 0.1180 | No |
| 15 | Ppard | 13226 | -0.024 | -0.0007 | No |
| 16 | Ccnd1 | 13886 | -0.043 | -0.0220 | No |
| 17 | Fzd7 | 14199 | -0.052 | -0.0273 | No |
| 18 | Cul1 | 15976 | -0.106 | -0.0865 | No |
| 19 | Notch1 | 16517 | -0.124 | -0.0905 | No |
| 20 | Fzd5 | 17738 | -0.169 | -0.1166 | No |
| 21 | St3gal6 | 19038 | -0.220 | -0.1383 | No |
| 22 | Jag1 | 19315 | -0.232 | -0.1148 | No |
| 23 | Skp1 | 20112 | -0.274 | -0.1070 | No |
| 24 | Notch3 | 20227 | -0.280 | -0.0693 | No |
| 25 | Kat2a | 20328 | -0.287 | -0.0301 | No |
| 26 | Rbx1 | 20448 | -0.291 | 0.0090 | No |
| 27 | Psenen | 21410 | -0.350 | 0.0214 | No |
| 28 | Maml2 | 22657 | -0.477 | 0.0410 | No |