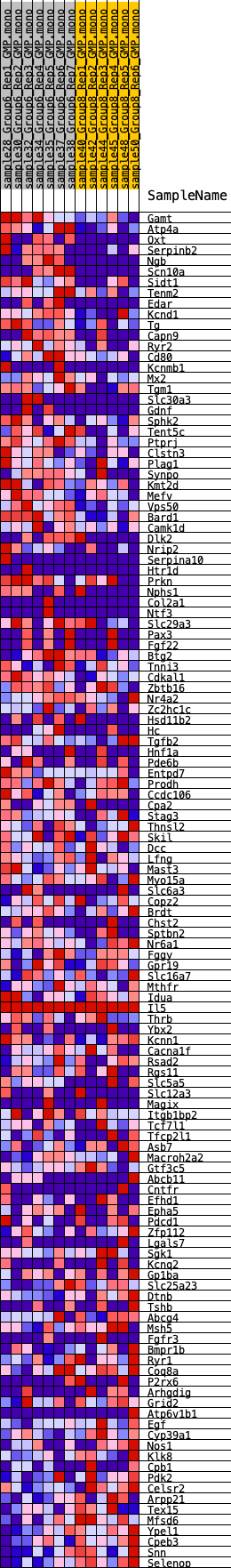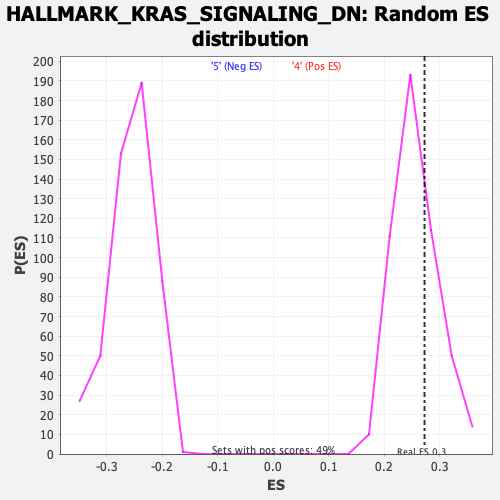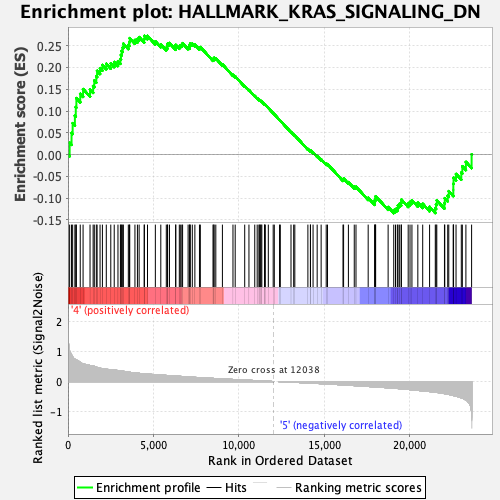
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group6_versus_Group8.GMP.mono_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | GMP.mono_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.27255228 |
| Normalized Enrichment Score (NES) | 1.0665271 |
| Nominal p-value | 0.29471543 |
| FDR q-value | 0.5569882 |
| FWER p-Value | 0.994 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gamt | 96 | 1.015 | 0.0271 | Yes |
| 2 | Atp4a | 207 | 0.890 | 0.0498 | Yes |
| 3 | Oxt | 272 | 0.816 | 0.0721 | Yes |
| 4 | Serpinb2 | 403 | 0.739 | 0.0892 | Yes |
| 5 | Ngb | 455 | 0.721 | 0.1092 | Yes |
| 6 | Scn10a | 491 | 0.713 | 0.1296 | Yes |
| 7 | Sidt1 | 716 | 0.631 | 0.1395 | Yes |
| 8 | Tenm2 | 882 | 0.583 | 0.1503 | Yes |
| 9 | Edar | 1289 | 0.530 | 0.1494 | Yes |
| 10 | Kcnd1 | 1470 | 0.516 | 0.1575 | Yes |
| 11 | Tg | 1538 | 0.501 | 0.1701 | Yes |
| 12 | Capn9 | 1656 | 0.478 | 0.1798 | Yes |
| 13 | Ryr2 | 1702 | 0.468 | 0.1922 | Yes |
| 14 | Cd80 | 1875 | 0.440 | 0.1984 | Yes |
| 15 | Kcnmb1 | 2013 | 0.425 | 0.2057 | Yes |
| 16 | Mx2 | 2240 | 0.405 | 0.2085 | Yes |
| 17 | Tgm1 | 2503 | 0.393 | 0.2094 | Yes |
| 18 | Slc30a3 | 2704 | 0.380 | 0.2125 | Yes |
| 19 | Gdnf | 2923 | 0.363 | 0.2144 | Yes |
| 20 | Sphk2 | 3066 | 0.356 | 0.2193 | Yes |
| 21 | Tent5c | 3098 | 0.353 | 0.2288 | Yes |
| 22 | Ptprj | 3137 | 0.349 | 0.2379 | Yes |
| 23 | Clstn3 | 3200 | 0.343 | 0.2458 | Yes |
| 24 | Plag1 | 3237 | 0.340 | 0.2547 | Yes |
| 25 | Synpo | 3529 | 0.312 | 0.2519 | Yes |
| 26 | Kmt2d | 3592 | 0.307 | 0.2587 | Yes |
| 27 | Mefv | 3612 | 0.306 | 0.2673 | Yes |
| 28 | Vps50 | 3916 | 0.285 | 0.2631 | Yes |
| 29 | Bard1 | 4062 | 0.277 | 0.2655 | Yes |
| 30 | Camk1d | 4161 | 0.271 | 0.2696 | Yes |
| 31 | Dlk2 | 4458 | 0.259 | 0.2650 | Yes |
| 32 | Nrip2 | 4468 | 0.259 | 0.2726 | Yes |
| 33 | Serpina10 | 4651 | 0.249 | 0.2724 | No |
| 34 | Htr1d | 5110 | 0.225 | 0.2599 | No |
| 35 | Prkn | 5429 | 0.218 | 0.2530 | No |
| 36 | Nphs1 | 5750 | 0.202 | 0.2456 | No |
| 37 | Col2a1 | 5815 | 0.199 | 0.2490 | No |
| 38 | Ntf3 | 5829 | 0.199 | 0.2546 | No |
| 39 | Slc29a3 | 5936 | 0.198 | 0.2561 | No |
| 40 | Pax3 | 6301 | 0.182 | 0.2462 | No |
| 41 | Fgf22 | 6302 | 0.182 | 0.2518 | No |
| 42 | Btg2 | 6512 | 0.174 | 0.2482 | No |
| 43 | Tnni3 | 6570 | 0.171 | 0.2511 | No |
| 44 | Cdkal1 | 6659 | 0.167 | 0.2524 | No |
| 45 | Zbtb16 | 6704 | 0.166 | 0.2557 | No |
| 46 | Nr4a2 | 7027 | 0.153 | 0.2467 | No |
| 47 | Zc2hc1c | 7118 | 0.150 | 0.2474 | No |
| 48 | Hsd11b2 | 7122 | 0.149 | 0.2519 | No |
| 49 | Hc | 7152 | 0.148 | 0.2552 | No |
| 50 | Tgfb2 | 7272 | 0.144 | 0.2545 | No |
| 51 | Hnf1a | 7422 | 0.138 | 0.2525 | No |
| 52 | Pde6b | 7691 | 0.129 | 0.2450 | No |
| 53 | Entpd7 | 7742 | 0.127 | 0.2468 | No |
| 54 | Prodh | 8484 | 0.103 | 0.2184 | No |
| 55 | Ccdc106 | 8535 | 0.102 | 0.2194 | No |
| 56 | Cpa2 | 8542 | 0.101 | 0.2223 | No |
| 57 | Stag3 | 8647 | 0.098 | 0.2208 | No |
| 58 | Thnsl2 | 9033 | 0.087 | 0.2071 | No |
| 59 | Skil | 9651 | 0.069 | 0.1830 | No |
| 60 | Dcc | 9781 | 0.065 | 0.1795 | No |
| 61 | Lfng | 10338 | 0.048 | 0.1573 | No |
| 62 | Mast3 | 10586 | 0.042 | 0.1481 | No |
| 63 | Myo15a | 10925 | 0.032 | 0.1347 | No |
| 64 | Slc6a3 | 11068 | 0.028 | 0.1295 | No |
| 65 | Copz2 | 11174 | 0.025 | 0.1258 | No |
| 66 | Brdt | 11193 | 0.025 | 0.1258 | No |
| 67 | Chst2 | 11252 | 0.022 | 0.1240 | No |
| 68 | Sptbn2 | 11293 | 0.022 | 0.1229 | No |
| 69 | Nr6a1 | 11339 | 0.020 | 0.1216 | No |
| 70 | Fggy | 11500 | 0.015 | 0.1153 | No |
| 71 | Gpr19 | 11501 | 0.015 | 0.1157 | No |
| 72 | Slc16a7 | 11542 | 0.014 | 0.1145 | No |
| 73 | Mthfr | 11719 | 0.009 | 0.1073 | No |
| 74 | Idua | 12004 | 0.001 | 0.0952 | No |
| 75 | Il5 | 12069 | 0.000 | 0.0925 | No |
| 76 | Thrb | 12375 | -0.002 | 0.0795 | No |
| 77 | Ybx2 | 12421 | -0.003 | 0.0777 | No |
| 78 | Kcnn1 | 13046 | -0.019 | 0.0518 | No |
| 79 | Cacna1f | 13203 | -0.024 | 0.0458 | No |
| 80 | Rsad2 | 13269 | -0.026 | 0.0439 | No |
| 81 | Rgs11 | 14034 | -0.048 | 0.0128 | No |
| 82 | Slc5a5 | 14182 | -0.052 | 0.0082 | No |
| 83 | Slc12a3 | 14185 | -0.052 | 0.0097 | No |
| 84 | Magix | 14337 | -0.056 | 0.0050 | No |
| 85 | Itgb1bp2 | 14577 | -0.063 | -0.0033 | No |
| 86 | Tcf7l1 | 14809 | -0.070 | -0.0110 | No |
| 87 | Tfcp2l1 | 15106 | -0.079 | -0.0211 | No |
| 88 | Asb7 | 15182 | -0.081 | -0.0218 | No |
| 89 | Macroh2a2 | 16088 | -0.110 | -0.0570 | No |
| 90 | Gtf3c5 | 16123 | -0.111 | -0.0550 | No |
| 91 | Abcb11 | 16404 | -0.120 | -0.0632 | No |
| 92 | Cntfr | 16747 | -0.133 | -0.0737 | No |
| 93 | Efhd1 | 16848 | -0.136 | -0.0738 | No |
| 94 | Epha5 | 17561 | -0.162 | -0.0991 | No |
| 95 | Pdcd1 | 17944 | -0.177 | -0.1099 | No |
| 96 | Zfp112 | 17948 | -0.177 | -0.1046 | No |
| 97 | Lgals7 | 17984 | -0.178 | -0.1006 | No |
| 98 | Sgk1 | 17998 | -0.179 | -0.0957 | No |
| 99 | Kcnq2 | 18728 | -0.207 | -0.1204 | No |
| 100 | Gp1ba | 19057 | -0.221 | -0.1276 | No |
| 101 | Slc25a23 | 19160 | -0.224 | -0.1250 | No |
| 102 | Dtnb | 19266 | -0.230 | -0.1224 | No |
| 103 | Tshb | 19302 | -0.231 | -0.1168 | No |
| 104 | Abcg4 | 19384 | -0.235 | -0.1130 | No |
| 105 | Msh5 | 19489 | -0.240 | -0.1101 | No |
| 106 | Fgfr3 | 19505 | -0.241 | -0.1033 | No |
| 107 | Bmpr1b | 19902 | -0.262 | -0.1121 | No |
| 108 | Ryr1 | 20009 | -0.267 | -0.1084 | No |
| 109 | Coq8a | 20121 | -0.275 | -0.1047 | No |
| 110 | P2rx6 | 20466 | -0.292 | -0.1104 | No |
| 111 | Arhgdig | 20751 | -0.309 | -0.1130 | No |
| 112 | Grid2 | 21154 | -0.332 | -0.1199 | No |
| 113 | Atp6v1b1 | 21487 | -0.354 | -0.1232 | No |
| 114 | Egf | 21532 | -0.356 | -0.1141 | No |
| 115 | Cyp39a1 | 21575 | -0.360 | -0.1049 | No |
| 116 | Nos1 | 22024 | -0.400 | -0.1117 | No |
| 117 | Klk8 | 22042 | -0.401 | -0.1001 | No |
| 118 | Cpb1 | 22215 | -0.421 | -0.0945 | No |
| 119 | Pdk2 | 22276 | -0.427 | -0.0839 | No |
| 120 | Celsr2 | 22542 | -0.460 | -0.0810 | No |
| 121 | Arpp21 | 22543 | -0.461 | -0.0669 | No |
| 122 | Tex15 | 22556 | -0.463 | -0.0532 | No |
| 123 | Mfsd6 | 22701 | -0.482 | -0.0445 | No |
| 124 | Ypel1 | 23009 | -0.537 | -0.0411 | No |
| 125 | Cpeb3 | 23070 | -0.549 | -0.0268 | No |
| 126 | Snn | 23278 | -0.611 | -0.0168 | No |
| 127 | Selenop | 23612 | -1.025 | 0.0005 | No |