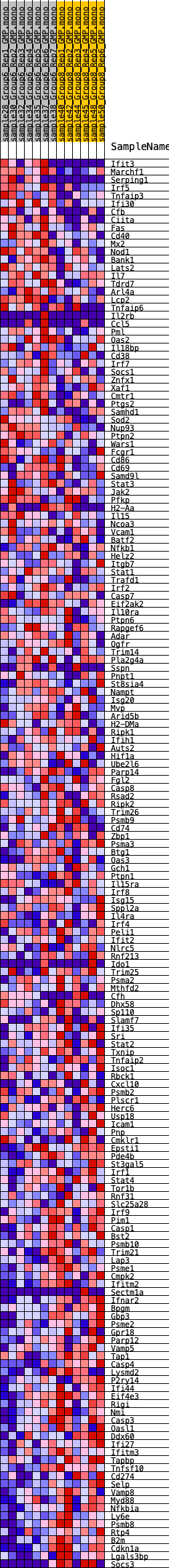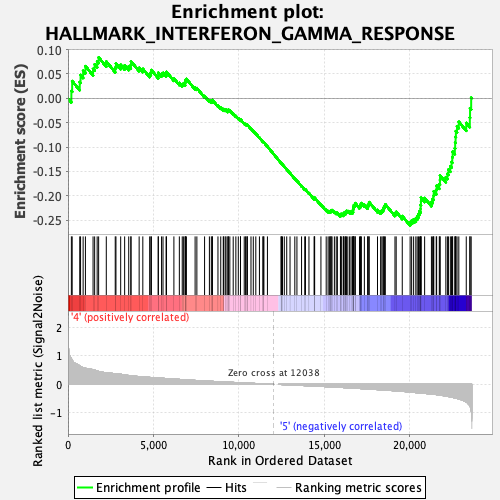
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group6_versus_Group8.GMP.mono_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | GMP.mono_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_INTERFERON_GAMMA_RESPONSE |
| Enrichment Score (ES) | -0.2610871 |
| Normalized Enrichment Score (NES) | -1.0699424 |
| Nominal p-value | 0.39615384 |
| FDR q-value | 0.70641583 |
| FWER p-Value | 0.994 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ifit3 | 197 | 0.902 | 0.0153 | No |
| 2 | Marchf1 | 244 | 0.843 | 0.0354 | No |
| 3 | Serping1 | 687 | 0.645 | 0.0335 | No |
| 4 | Irf5 | 735 | 0.626 | 0.0479 | No |
| 5 | Tnfaip3 | 885 | 0.582 | 0.0568 | No |
| 6 | Ifi30 | 1019 | 0.567 | 0.0660 | No |
| 7 | Cfb | 1466 | 0.517 | 0.0606 | No |
| 8 | Ciita | 1559 | 0.498 | 0.0697 | No |
| 9 | Fas | 1706 | 0.468 | 0.0758 | No |
| 10 | Cd40 | 1792 | 0.451 | 0.0840 | No |
| 11 | Mx2 | 2240 | 0.405 | 0.0755 | No |
| 12 | Nod1 | 2763 | 0.378 | 0.0632 | No |
| 13 | Bank1 | 2805 | 0.375 | 0.0713 | No |
| 14 | Lats2 | 3082 | 0.354 | 0.0688 | No |
| 15 | Il7 | 3314 | 0.332 | 0.0677 | No |
| 16 | Tdrd7 | 3552 | 0.311 | 0.0657 | No |
| 17 | Arl4a | 3676 | 0.301 | 0.0684 | No |
| 18 | Lcp2 | 3687 | 0.301 | 0.0758 | No |
| 19 | Tnfaip6 | 4162 | 0.271 | 0.0628 | No |
| 20 | Il2rb | 4377 | 0.261 | 0.0605 | No |
| 21 | Ccl5 | 4778 | 0.244 | 0.0498 | No |
| 22 | Pml | 4846 | 0.240 | 0.0532 | No |
| 23 | Oas2 | 4880 | 0.237 | 0.0581 | No |
| 24 | Il18bp | 5280 | 0.223 | 0.0469 | No |
| 25 | Cd38 | 5282 | 0.223 | 0.0527 | No |
| 26 | Irf7 | 5473 | 0.216 | 0.0503 | No |
| 27 | Socs1 | 5558 | 0.212 | 0.0523 | No |
| 28 | Znfx1 | 5738 | 0.203 | 0.0499 | No |
| 29 | Xaf1 | 5763 | 0.201 | 0.0542 | No |
| 30 | Cmtr1 | 6195 | 0.187 | 0.0407 | No |
| 31 | Ptgs2 | 6514 | 0.173 | 0.0317 | No |
| 32 | Samhd1 | 6683 | 0.167 | 0.0289 | No |
| 33 | Sod2 | 6749 | 0.165 | 0.0305 | No |
| 34 | Nup93 | 6852 | 0.161 | 0.0303 | No |
| 35 | Ptpn2 | 6857 | 0.161 | 0.0344 | No |
| 36 | Wars1 | 6876 | 0.160 | 0.0378 | No |
| 37 | Fcgr1 | 6927 | 0.158 | 0.0398 | No |
| 38 | Cd86 | 7436 | 0.138 | 0.0218 | No |
| 39 | Cd69 | 7539 | 0.134 | 0.0209 | No |
| 40 | Samd9l | 7990 | 0.118 | 0.0048 | No |
| 41 | Stat3 | 8272 | 0.110 | -0.0043 | No |
| 42 | Jak2 | 8385 | 0.107 | -0.0062 | No |
| 43 | Pfkp | 8439 | 0.105 | -0.0057 | No |
| 44 | H2-Aa | 8446 | 0.105 | -0.0032 | No |
| 45 | Il15 | 8773 | 0.094 | -0.0147 | No |
| 46 | Ncoa3 | 8936 | 0.089 | -0.0193 | No |
| 47 | Vcam1 | 9083 | 0.085 | -0.0232 | No |
| 48 | Batf2 | 9105 | 0.085 | -0.0219 | No |
| 49 | Nfkb1 | 9196 | 0.082 | -0.0236 | No |
| 50 | Helz2 | 9245 | 0.081 | -0.0235 | No |
| 51 | Itgb7 | 9356 | 0.077 | -0.0262 | No |
| 52 | Stat1 | 9367 | 0.077 | -0.0246 | No |
| 53 | Trafd1 | 9381 | 0.076 | -0.0231 | No |
| 54 | Irf2 | 9467 | 0.074 | -0.0248 | No |
| 55 | Casp7 | 9665 | 0.068 | -0.0314 | No |
| 56 | Eif2ak2 | 9820 | 0.064 | -0.0363 | No |
| 57 | Il10ra | 9963 | 0.060 | -0.0408 | No |
| 58 | Ptpn6 | 10091 | 0.056 | -0.0448 | No |
| 59 | Rapgef6 | 10092 | 0.056 | -0.0433 | No |
| 60 | Adar | 10310 | 0.049 | -0.0513 | No |
| 61 | Ogfr | 10391 | 0.047 | -0.0535 | No |
| 62 | Trim14 | 10414 | 0.046 | -0.0532 | No |
| 63 | Pla2g4a | 10487 | 0.045 | -0.0551 | No |
| 64 | Sspn | 10490 | 0.045 | -0.0540 | No |
| 65 | Pnpt1 | 10705 | 0.039 | -0.0621 | No |
| 66 | St8sia4 | 10840 | 0.035 | -0.0669 | No |
| 67 | Nampt | 10987 | 0.031 | -0.0723 | No |
| 68 | Isg20 | 11199 | 0.024 | -0.0807 | No |
| 69 | Mvp | 11399 | 0.018 | -0.0887 | No |
| 70 | Arid5b | 11428 | 0.017 | -0.0895 | No |
| 71 | H2-DMa | 11461 | 0.016 | -0.0904 | No |
| 72 | Ripk1 | 11667 | 0.010 | -0.0989 | No |
| 73 | Ifih1 | 12463 | -0.004 | -0.1327 | No |
| 74 | Auts2 | 12481 | -0.004 | -0.1333 | No |
| 75 | Hif1a | 12503 | -0.005 | -0.1341 | No |
| 76 | Ube2l6 | 12548 | -0.006 | -0.1358 | No |
| 77 | Parp14 | 12654 | -0.008 | -0.1400 | No |
| 78 | Fgl2 | 12796 | -0.012 | -0.1457 | No |
| 79 | Casp8 | 12989 | -0.018 | -0.1534 | No |
| 80 | Rsad2 | 13269 | -0.026 | -0.1647 | No |
| 81 | Ripk2 | 13393 | -0.029 | -0.1692 | No |
| 82 | Trim26 | 13664 | -0.037 | -0.1797 | No |
| 83 | Psmb9 | 13851 | -0.042 | -0.1865 | No |
| 84 | Cd74 | 13892 | -0.043 | -0.1871 | No |
| 85 | Zbp1 | 14096 | -0.050 | -0.1944 | No |
| 86 | Psma3 | 14407 | -0.058 | -0.2061 | No |
| 87 | Btg1 | 14408 | -0.058 | -0.2046 | No |
| 88 | Oas3 | 14430 | -0.058 | -0.2040 | No |
| 89 | Gch1 | 14797 | -0.069 | -0.2178 | No |
| 90 | Ptpn1 | 15105 | -0.079 | -0.2288 | No |
| 91 | Il15ra | 15216 | -0.083 | -0.2313 | No |
| 92 | Irf8 | 15289 | -0.086 | -0.2321 | No |
| 93 | Isg15 | 15336 | -0.087 | -0.2318 | No |
| 94 | Sppl2a | 15383 | -0.089 | -0.2314 | No |
| 95 | Il4ra | 15393 | -0.089 | -0.2295 | No |
| 96 | Irf4 | 15447 | -0.090 | -0.2294 | No |
| 97 | Peli1 | 15584 | -0.094 | -0.2327 | No |
| 98 | Ifit2 | 15691 | -0.097 | -0.2347 | No |
| 99 | Nlrc5 | 15759 | -0.099 | -0.2349 | No |
| 100 | Rnf213 | 15938 | -0.105 | -0.2398 | No |
| 101 | Ido1 | 15950 | -0.105 | -0.2375 | No |
| 102 | Trim25 | 16009 | -0.107 | -0.2371 | No |
| 103 | Psma2 | 16107 | -0.110 | -0.2384 | No |
| 104 | Mthfd2 | 16122 | -0.111 | -0.2361 | No |
| 105 | Cfh | 16154 | -0.112 | -0.2345 | No |
| 106 | Dhx58 | 16241 | -0.115 | -0.2351 | No |
| 107 | Sp110 | 16248 | -0.115 | -0.2323 | No |
| 108 | Slamf7 | 16288 | -0.117 | -0.2310 | No |
| 109 | Ifi35 | 16369 | -0.119 | -0.2312 | No |
| 110 | Sri | 16475 | -0.123 | -0.2325 | No |
| 111 | Stat2 | 16559 | -0.127 | -0.2327 | No |
| 112 | Txnip | 16646 | -0.130 | -0.2330 | No |
| 113 | Tnfaip2 | 16675 | -0.131 | -0.2307 | No |
| 114 | Isoc1 | 16680 | -0.131 | -0.2275 | No |
| 115 | Rbck1 | 16691 | -0.131 | -0.2244 | No |
| 116 | Cxcl10 | 16716 | -0.132 | -0.2220 | No |
| 117 | Psmb2 | 16728 | -0.133 | -0.2190 | No |
| 118 | Plscr1 | 16799 | -0.135 | -0.2185 | No |
| 119 | Herc6 | 16802 | -0.135 | -0.2150 | No |
| 120 | Usp18 | 17055 | -0.144 | -0.2220 | No |
| 121 | Icam1 | 17106 | -0.146 | -0.2203 | No |
| 122 | Pnp | 17124 | -0.146 | -0.2172 | No |
| 123 | Cmklr1 | 17163 | -0.148 | -0.2149 | No |
| 124 | Epsti1 | 17338 | -0.154 | -0.2183 | No |
| 125 | Pde4b | 17530 | -0.161 | -0.2222 | No |
| 126 | St3gal5 | 17549 | -0.162 | -0.2187 | No |
| 127 | Irf1 | 17602 | -0.164 | -0.2166 | No |
| 128 | Stat4 | 17618 | -0.165 | -0.2130 | No |
| 129 | Tor1b | 18106 | -0.182 | -0.2290 | No |
| 130 | Rnf31 | 18289 | -0.189 | -0.2318 | No |
| 131 | Slc25a28 | 18363 | -0.191 | -0.2299 | No |
| 132 | Irf9 | 18431 | -0.195 | -0.2276 | No |
| 133 | Pim1 | 18467 | -0.196 | -0.2239 | No |
| 134 | Casp1 | 18513 | -0.198 | -0.2207 | No |
| 135 | Bst2 | 18564 | -0.200 | -0.2175 | No |
| 136 | Psmb10 | 19129 | -0.223 | -0.2357 | No |
| 137 | Trim21 | 19199 | -0.226 | -0.2327 | No |
| 138 | Lap3 | 19555 | -0.243 | -0.2415 | No |
| 139 | Psme1 | 20015 | -0.268 | -0.2541 | Yes |
| 140 | Cmpk2 | 20104 | -0.274 | -0.2506 | Yes |
| 141 | Ifitm2 | 20220 | -0.280 | -0.2482 | Yes |
| 142 | Sectm1a | 20349 | -0.288 | -0.2461 | Yes |
| 143 | Ifnar2 | 20428 | -0.290 | -0.2418 | Yes |
| 144 | Bpgm | 20503 | -0.294 | -0.2373 | Yes |
| 145 | Gbp3 | 20556 | -0.298 | -0.2317 | Yes |
| 146 | Psme2 | 20627 | -0.302 | -0.2267 | Yes |
| 147 | Gpr18 | 20630 | -0.302 | -0.2189 | Yes |
| 148 | Parp12 | 20649 | -0.303 | -0.2117 | Yes |
| 149 | Vamp5 | 20651 | -0.303 | -0.2038 | Yes |
| 150 | Tap1 | 20864 | -0.317 | -0.2045 | Yes |
| 151 | Casp4 | 21261 | -0.340 | -0.2125 | Yes |
| 152 | Lysmd2 | 21332 | -0.344 | -0.2064 | Yes |
| 153 | P2ry14 | 21387 | -0.349 | -0.1996 | Yes |
| 154 | Ifi44 | 21397 | -0.350 | -0.1908 | Yes |
| 155 | Eif4e3 | 21542 | -0.357 | -0.1876 | Yes |
| 156 | Rigi | 21566 | -0.359 | -0.1791 | Yes |
| 157 | Nmi | 21722 | -0.371 | -0.1760 | Yes |
| 158 | Casp3 | 21760 | -0.375 | -0.1677 | Yes |
| 159 | Oasl1 | 21765 | -0.375 | -0.1581 | Yes |
| 160 | Ddx60 | 22105 | -0.408 | -0.1618 | Yes |
| 161 | Ifi27 | 22197 | -0.419 | -0.1547 | Yes |
| 162 | Ifitm3 | 22256 | -0.425 | -0.1460 | Yes |
| 163 | Tapbp | 22368 | -0.437 | -0.1393 | Yes |
| 164 | Tnfsf10 | 22439 | -0.446 | -0.1306 | Yes |
| 165 | Cd274 | 22482 | -0.452 | -0.1205 | Yes |
| 166 | Selp | 22503 | -0.455 | -0.1094 | Yes |
| 167 | Vamp8 | 22632 | -0.474 | -0.1024 | Yes |
| 168 | Myd88 | 22650 | -0.476 | -0.0907 | Yes |
| 169 | Nfkbia | 22674 | -0.479 | -0.0791 | Yes |
| 170 | Ly6e | 22692 | -0.481 | -0.0672 | Yes |
| 171 | Psmb8 | 22759 | -0.492 | -0.0571 | Yes |
| 172 | Rtp4 | 22864 | -0.509 | -0.0482 | Yes |
| 173 | B2m | 23299 | -0.618 | -0.0504 | Yes |
| 174 | Cdkn1a | 23508 | -0.752 | -0.0396 | Yes |
| 175 | Lgals3bp | 23521 | -0.763 | -0.0201 | Yes |
| 176 | Socs3 | 23587 | -0.931 | 0.0016 | Yes |