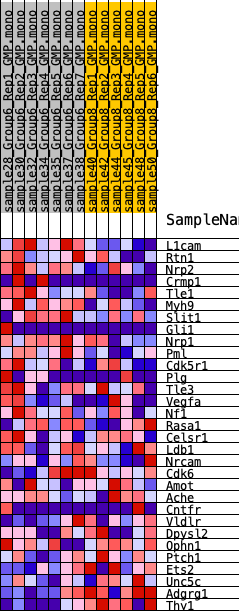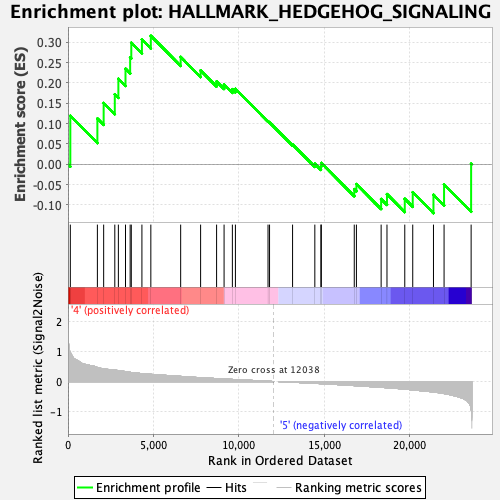
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group6_versus_Group8.GMP.mono_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | GMP.mono_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_HEDGEHOG_SIGNALING |
| Enrichment Score (ES) | 0.31578952 |
| Normalized Enrichment Score (NES) | 1.0174619 |
| Nominal p-value | 0.43887776 |
| FDR q-value | 0.5372623 |
| FWER p-Value | 0.998 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | L1cam | 136 | 0.966 | 0.1193 | Yes |
| 2 | Rtn1 | 1719 | 0.465 | 0.1124 | Yes |
| 3 | Nrp2 | 2086 | 0.415 | 0.1507 | Yes |
| 4 | Crmp1 | 2738 | 0.379 | 0.1721 | Yes |
| 5 | Tle1 | 2947 | 0.363 | 0.2103 | Yes |
| 6 | Myh9 | 3364 | 0.327 | 0.2350 | Yes |
| 7 | Slit1 | 3636 | 0.304 | 0.2629 | Yes |
| 8 | Gli1 | 3703 | 0.300 | 0.2990 | Yes |
| 9 | Nrp1 | 4324 | 0.264 | 0.3068 | Yes |
| 10 | Pml | 4846 | 0.240 | 0.3158 | Yes |
| 11 | Cdk5r1 | 6591 | 0.170 | 0.2639 | No |
| 12 | Plg | 7758 | 0.126 | 0.2309 | No |
| 13 | Tle3 | 8693 | 0.096 | 0.2038 | No |
| 14 | Vegfa | 9131 | 0.084 | 0.1961 | No |
| 15 | Nf1 | 9616 | 0.070 | 0.1846 | No |
| 16 | Rasa1 | 9794 | 0.064 | 0.1855 | No |
| 17 | Celsr1 | 11702 | 0.009 | 0.1059 | No |
| 18 | Ldb1 | 11791 | 0.007 | 0.1030 | No |
| 19 | Nrcam | 13138 | -0.022 | 0.0488 | No |
| 20 | Cdk6 | 14440 | -0.059 | 0.0013 | No |
| 21 | Amot | 14795 | -0.069 | -0.0048 | No |
| 22 | Ache | 14820 | -0.070 | 0.0033 | No |
| 23 | Cntfr | 16747 | -0.133 | -0.0611 | No |
| 24 | Vldlr | 16873 | -0.137 | -0.0487 | No |
| 25 | Dpysl2 | 18319 | -0.190 | -0.0854 | No |
| 26 | Ophn1 | 18661 | -0.204 | -0.0734 | No |
| 27 | Ptch1 | 19702 | -0.251 | -0.0849 | No |
| 28 | Ets2 | 20168 | -0.277 | -0.0687 | No |
| 29 | Unc5c | 21381 | -0.348 | -0.0750 | No |
| 30 | Adgrg1 | 22001 | -0.398 | -0.0497 | No |
| 31 | Thy1 | 23584 | -0.914 | 0.0017 | No |