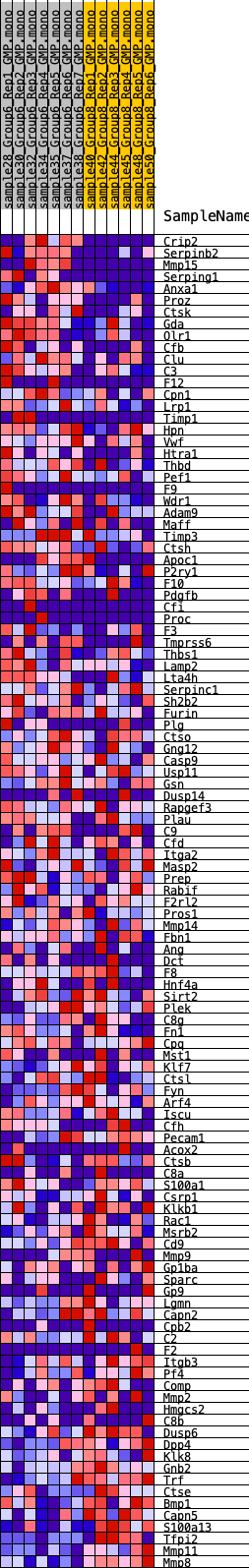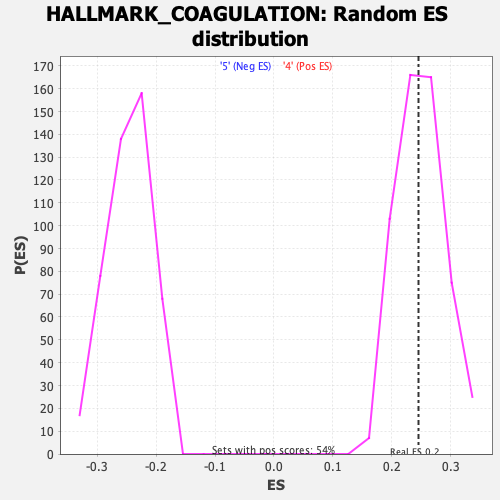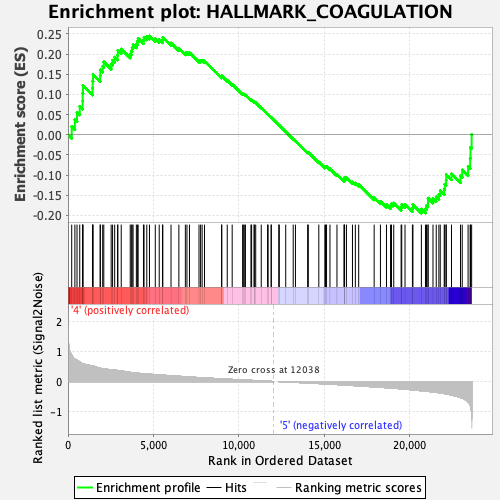
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group6_versus_Group8.GMP.mono_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | GMP.mono_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | 0.24521491 |
| Normalized Enrichment Score (NES) | 0.98301744 |
| Nominal p-value | 0.51756006 |
| FDR q-value | 0.577396 |
| FWER p-Value | 0.999 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Crip2 | 217 | 0.876 | 0.0203 | Yes |
| 2 | Serpinb2 | 403 | 0.739 | 0.0373 | Yes |
| 3 | Mmp15 | 528 | 0.703 | 0.0556 | Yes |
| 4 | Serping1 | 687 | 0.645 | 0.0706 | Yes |
| 5 | Anxa1 | 856 | 0.589 | 0.0833 | Yes |
| 6 | Proz | 859 | 0.588 | 0.1030 | Yes |
| 7 | Ctsk | 877 | 0.586 | 0.1220 | Yes |
| 8 | Gda | 1433 | 0.520 | 0.1159 | Yes |
| 9 | Olr1 | 1445 | 0.519 | 0.1329 | Yes |
| 10 | Cfb | 1466 | 0.517 | 0.1495 | Yes |
| 11 | Clu | 1883 | 0.439 | 0.1466 | Yes |
| 12 | C3 | 1890 | 0.439 | 0.1611 | Yes |
| 13 | F12 | 2029 | 0.425 | 0.1695 | Yes |
| 14 | Cpn1 | 2094 | 0.414 | 0.1807 | Yes |
| 15 | Lrp1 | 2525 | 0.390 | 0.1755 | Yes |
| 16 | Timp1 | 2608 | 0.382 | 0.1849 | Yes |
| 17 | Hpn | 2731 | 0.379 | 0.1925 | Yes |
| 18 | Vwf | 2901 | 0.365 | 0.1976 | Yes |
| 19 | Htra1 | 2917 | 0.363 | 0.2091 | Yes |
| 20 | Thbd | 3114 | 0.351 | 0.2126 | Yes |
| 21 | Pef1 | 3640 | 0.304 | 0.2005 | Yes |
| 22 | F9 | 3704 | 0.300 | 0.2079 | Yes |
| 23 | Wdr1 | 3753 | 0.297 | 0.2159 | Yes |
| 24 | Adam9 | 3808 | 0.293 | 0.2235 | Yes |
| 25 | Maff | 3999 | 0.281 | 0.2248 | Yes |
| 26 | Timp3 | 4059 | 0.277 | 0.2317 | Yes |
| 27 | Ctsh | 4103 | 0.275 | 0.2391 | Yes |
| 28 | Apoc1 | 4415 | 0.261 | 0.2346 | Yes |
| 29 | P2ry1 | 4448 | 0.260 | 0.2420 | Yes |
| 30 | F10 | 4610 | 0.250 | 0.2436 | Yes |
| 31 | Pdgfb | 4766 | 0.245 | 0.2452 | Yes |
| 32 | Cfi | 5106 | 0.225 | 0.2384 | No |
| 33 | Proc | 5336 | 0.220 | 0.2360 | No |
| 34 | F3 | 5527 | 0.213 | 0.2351 | No |
| 35 | Tmprss6 | 5548 | 0.213 | 0.2415 | No |
| 36 | Thbs1 | 6027 | 0.194 | 0.2277 | No |
| 37 | Lamp2 | 6486 | 0.175 | 0.2141 | No |
| 38 | Lta4h | 6865 | 0.160 | 0.2034 | No |
| 39 | Serpinc1 | 6959 | 0.156 | 0.2047 | No |
| 40 | Sh2b2 | 7106 | 0.150 | 0.2035 | No |
| 41 | Furin | 7664 | 0.129 | 0.1842 | No |
| 42 | Plg | 7758 | 0.126 | 0.1845 | No |
| 43 | Ctso | 7847 | 0.124 | 0.1849 | No |
| 44 | Gng12 | 7986 | 0.119 | 0.1830 | No |
| 45 | Casp9 | 8986 | 0.088 | 0.1435 | No |
| 46 | Usp11 | 8996 | 0.088 | 0.1460 | No |
| 47 | Gsn | 9316 | 0.079 | 0.1351 | No |
| 48 | Dusp14 | 9605 | 0.070 | 0.1252 | No |
| 49 | Rapgef3 | 10217 | 0.052 | 0.1010 | No |
| 50 | Plau | 10255 | 0.051 | 0.1011 | No |
| 51 | C9 | 10312 | 0.049 | 0.1004 | No |
| 52 | Cfd | 10380 | 0.047 | 0.0991 | No |
| 53 | Itga2 | 10703 | 0.039 | 0.0867 | No |
| 54 | Masp2 | 10742 | 0.038 | 0.0864 | No |
| 55 | Prep | 10889 | 0.033 | 0.0813 | No |
| 56 | Rabif | 10898 | 0.033 | 0.0821 | No |
| 57 | F2rl2 | 10981 | 0.031 | 0.0796 | No |
| 58 | Pros1 | 11305 | 0.021 | 0.0666 | No |
| 59 | Mmp14 | 11676 | 0.010 | 0.0512 | No |
| 60 | Fbn1 | 11707 | 0.009 | 0.0502 | No |
| 61 | Ang | 11886 | 0.004 | 0.0428 | No |
| 62 | Dct | 11892 | 0.004 | 0.0427 | No |
| 63 | F8 | 12331 | -0.000 | 0.0241 | No |
| 64 | Hnf4a | 12361 | -0.001 | 0.0229 | No |
| 65 | Sirt2 | 12737 | -0.011 | 0.0073 | No |
| 66 | Plek | 13175 | -0.023 | -0.0105 | No |
| 67 | C8g | 13306 | -0.027 | -0.0151 | No |
| 68 | Fn1 | 14020 | -0.047 | -0.0439 | No |
| 69 | Cpq | 14058 | -0.048 | -0.0438 | No |
| 70 | Mst1 | 14673 | -0.065 | -0.0677 | No |
| 71 | Klf7 | 15027 | -0.077 | -0.0802 | No |
| 72 | Ctsl | 15065 | -0.078 | -0.0791 | No |
| 73 | Fyn | 15125 | -0.080 | -0.0789 | No |
| 74 | Arf4 | 15326 | -0.087 | -0.0845 | No |
| 75 | Iscu | 15735 | -0.098 | -0.0986 | No |
| 76 | Cfh | 16154 | -0.112 | -0.1126 | No |
| 77 | Pecam1 | 16159 | -0.112 | -0.1090 | No |
| 78 | Acox2 | 16179 | -0.113 | -0.1060 | No |
| 79 | Ctsb | 16290 | -0.117 | -0.1067 | No |
| 80 | C8a | 16642 | -0.130 | -0.1173 | No |
| 81 | S100a1 | 16810 | -0.135 | -0.1198 | No |
| 82 | Csrp1 | 17006 | -0.142 | -0.1234 | No |
| 83 | Klkb1 | 17911 | -0.176 | -0.1559 | No |
| 84 | Rac1 | 18278 | -0.188 | -0.1651 | No |
| 85 | Msrb2 | 18632 | -0.203 | -0.1733 | No |
| 86 | Cd9 | 18870 | -0.212 | -0.1762 | No |
| 87 | Mmp9 | 18925 | -0.215 | -0.1713 | No |
| 88 | Gp1ba | 19057 | -0.221 | -0.1694 | No |
| 89 | Sparc | 19488 | -0.240 | -0.1796 | No |
| 90 | Gp9 | 19525 | -0.241 | -0.1731 | No |
| 91 | Lgmn | 19716 | -0.252 | -0.1727 | No |
| 92 | Capn2 | 20149 | -0.276 | -0.1817 | No |
| 93 | Cpb2 | 20180 | -0.278 | -0.1737 | No |
| 94 | C2 | 20673 | -0.304 | -0.1843 | No |
| 95 | F2 | 20913 | -0.319 | -0.1838 | No |
| 96 | Itgb3 | 20975 | -0.321 | -0.1756 | No |
| 97 | Pf4 | 21062 | -0.327 | -0.1682 | No |
| 98 | Comp | 21064 | -0.327 | -0.1573 | No |
| 99 | Mmp2 | 21353 | -0.346 | -0.1579 | No |
| 100 | Hmgcs2 | 21543 | -0.357 | -0.1539 | No |
| 101 | C8b | 21697 | -0.369 | -0.1480 | No |
| 102 | Dusp6 | 21777 | -0.376 | -0.1387 | No |
| 103 | Dpp4 | 22010 | -0.398 | -0.1352 | No |
| 104 | Klk8 | 22042 | -0.401 | -0.1230 | No |
| 105 | Gnb2 | 22121 | -0.410 | -0.1125 | No |
| 106 | Trf | 22128 | -0.411 | -0.0989 | No |
| 107 | Ctse | 22435 | -0.445 | -0.0969 | No |
| 108 | Bmp1 | 22962 | -0.528 | -0.1015 | No |
| 109 | Capn5 | 23063 | -0.547 | -0.0874 | No |
| 110 | S100a13 | 23406 | -0.674 | -0.0792 | No |
| 111 | Tfpi2 | 23527 | -0.776 | -0.0582 | No |
| 112 | Mmp11 | 23547 | -0.819 | -0.0315 | No |
| 113 | Mmp8 | 23614 | -1.032 | 0.0004 | No |