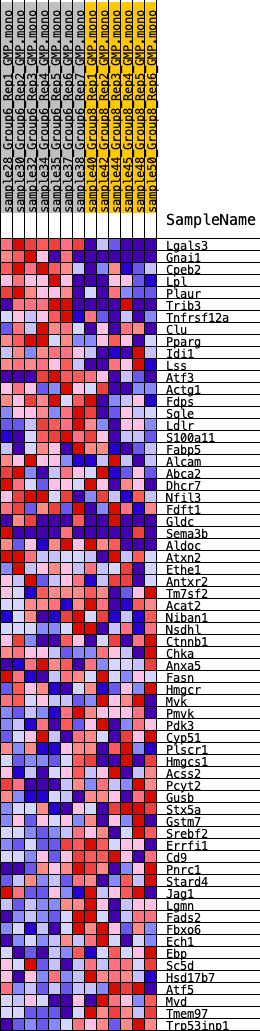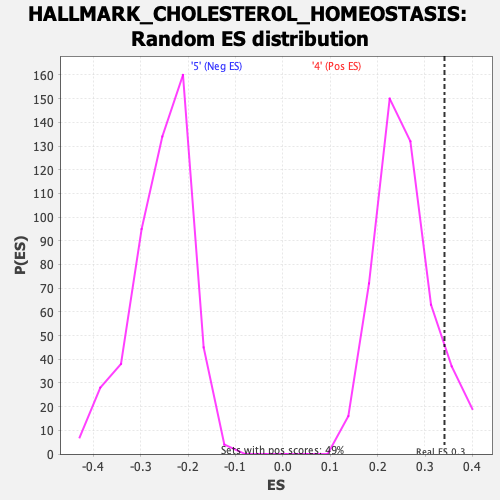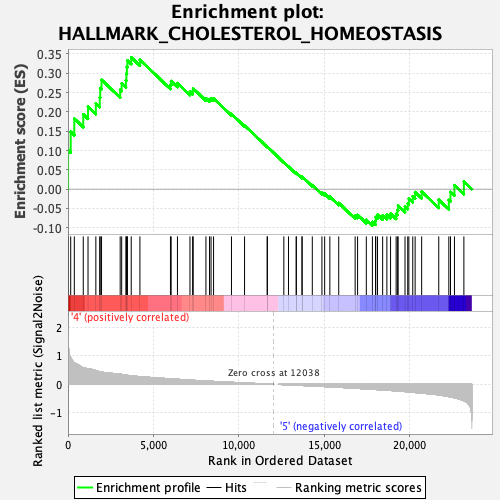
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group6_versus_Group8.GMP.mono_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | GMP.mono_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | 0.34111366 |
| Normalized Enrichment Score (NES) | 1.32726 |
| Nominal p-value | 0.10429448 |
| FDR q-value | 0.5493782 |
| FWER p-Value | 0.727 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Lgals3 | 3 | 1.760 | 0.1010 | Yes |
| 2 | Gnai1 | 158 | 0.940 | 0.1484 | Yes |
| 3 | Cpeb2 | 369 | 0.750 | 0.1826 | Yes |
| 4 | Lpl | 893 | 0.581 | 0.1937 | Yes |
| 5 | Plaur | 1171 | 0.548 | 0.2135 | Yes |
| 6 | Trib3 | 1629 | 0.483 | 0.2218 | Yes |
| 7 | Tnfrsf12a | 1857 | 0.444 | 0.2376 | Yes |
| 8 | Clu | 1883 | 0.439 | 0.2618 | Yes |
| 9 | Pparg | 1962 | 0.431 | 0.2832 | Yes |
| 10 | Idi1 | 3052 | 0.357 | 0.2575 | Yes |
| 11 | Lss | 3140 | 0.348 | 0.2738 | Yes |
| 12 | Atf3 | 3388 | 0.324 | 0.2819 | Yes |
| 13 | Actg1 | 3417 | 0.322 | 0.2992 | Yes |
| 14 | Fdps | 3444 | 0.319 | 0.3164 | Yes |
| 15 | Sqle | 3477 | 0.316 | 0.3333 | Yes |
| 16 | Ldlr | 3699 | 0.300 | 0.3411 | Yes |
| 17 | S100a11 | 4207 | 0.268 | 0.3350 | No |
| 18 | Fabp5 | 5994 | 0.196 | 0.2704 | No |
| 19 | Alcam | 6034 | 0.194 | 0.2799 | No |
| 20 | Abca2 | 6403 | 0.178 | 0.2745 | No |
| 21 | Dhcr7 | 7135 | 0.149 | 0.2520 | No |
| 22 | Nfil3 | 7291 | 0.143 | 0.2536 | No |
| 23 | Fdft1 | 7324 | 0.142 | 0.2604 | No |
| 24 | Gldc | 8069 | 0.116 | 0.2355 | No |
| 25 | Sema3b | 8279 | 0.110 | 0.2329 | No |
| 26 | Aldoc | 8369 | 0.107 | 0.2353 | No |
| 27 | Atxn2 | 8516 | 0.102 | 0.2350 | No |
| 28 | Ethe1 | 9563 | 0.071 | 0.1946 | No |
| 29 | Antxr2 | 10329 | 0.049 | 0.1650 | No |
| 30 | Tm7sf2 | 11652 | 0.011 | 0.1095 | No |
| 31 | Acat2 | 11664 | 0.010 | 0.1096 | No |
| 32 | Niban1 | 12627 | -0.008 | 0.0692 | No |
| 33 | Nsdhl | 12899 | -0.015 | 0.0586 | No |
| 34 | Ctnnb1 | 13353 | -0.028 | 0.0409 | No |
| 35 | Chka | 13355 | -0.028 | 0.0425 | No |
| 36 | Anxa5 | 13692 | -0.038 | 0.0304 | No |
| 37 | Fasn | 13694 | -0.038 | 0.0325 | No |
| 38 | Hmgcr | 14292 | -0.055 | 0.0103 | No |
| 39 | Mvk | 14856 | -0.071 | -0.0095 | No |
| 40 | Pmvk | 15015 | -0.076 | -0.0118 | No |
| 41 | Pdk3 | 15319 | -0.087 | -0.0197 | No |
| 42 | Cyp51 | 15838 | -0.102 | -0.0358 | No |
| 43 | Plscr1 | 16799 | -0.135 | -0.0689 | No |
| 44 | Hmgcs1 | 16942 | -0.140 | -0.0669 | No |
| 45 | Acss2 | 17452 | -0.158 | -0.0794 | No |
| 46 | Pcyt2 | 17808 | -0.172 | -0.0846 | No |
| 47 | Gusb | 17992 | -0.179 | -0.0821 | No |
| 48 | Stx5a | 18004 | -0.179 | -0.0723 | No |
| 49 | Gstm7 | 18108 | -0.182 | -0.0662 | No |
| 50 | Srebf2 | 18408 | -0.194 | -0.0678 | No |
| 51 | Errfi1 | 18655 | -0.204 | -0.0665 | No |
| 52 | Cd9 | 18870 | -0.212 | -0.0634 | No |
| 53 | Pnrc1 | 19196 | -0.226 | -0.0642 | No |
| 54 | Stard4 | 19269 | -0.230 | -0.0541 | No |
| 55 | Jag1 | 19315 | -0.232 | -0.0427 | No |
| 56 | Lgmn | 19716 | -0.252 | -0.0452 | No |
| 57 | Fads2 | 19875 | -0.260 | -0.0370 | No |
| 58 | Fbxo6 | 19941 | -0.264 | -0.0246 | No |
| 59 | Ech1 | 20169 | -0.277 | -0.0183 | No |
| 60 | Ebp | 20308 | -0.285 | -0.0078 | No |
| 61 | Sc5d | 20690 | -0.305 | -0.0064 | No |
| 62 | Hsd17b7 | 21690 | -0.369 | -0.0276 | No |
| 63 | Atf5 | 22279 | -0.427 | -0.0281 | No |
| 64 | Mvd | 22377 | -0.438 | -0.0071 | No |
| 65 | Tmem97 | 22608 | -0.471 | 0.0102 | No |
| 66 | Trp53inp1 | 23160 | -0.573 | 0.0197 | No |