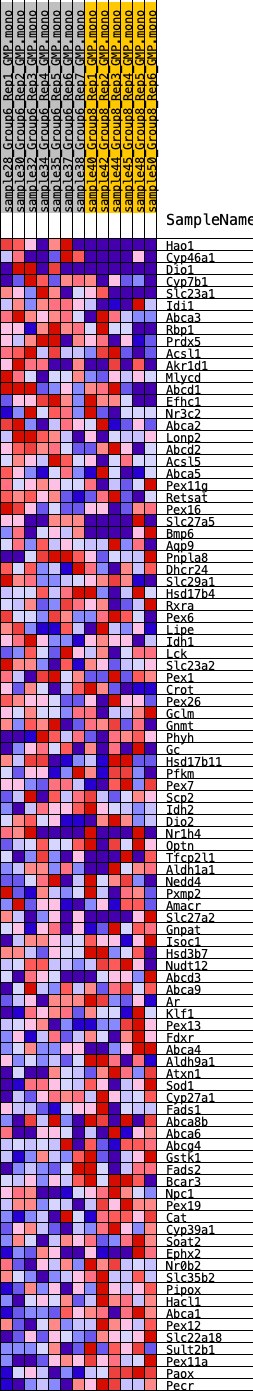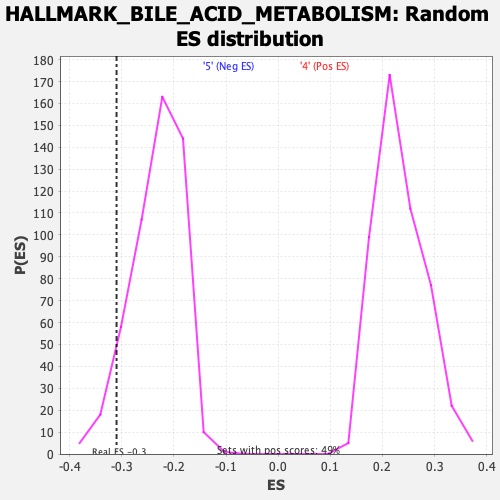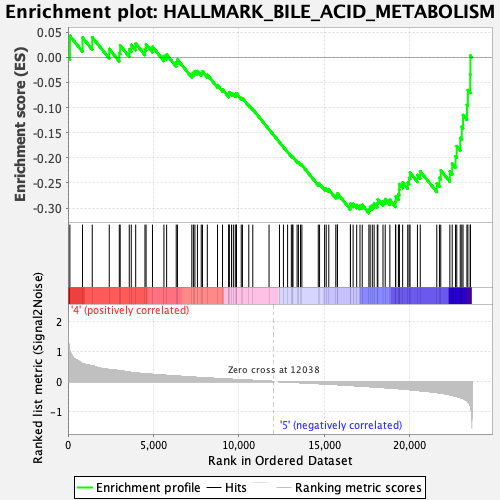
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group6_versus_Group8.GMP.mono_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | GMP.mono_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.31009576 |
| Normalized Enrichment Score (NES) | -1.3247268 |
| Nominal p-value | 0.05928854 |
| FDR q-value | 0.474731 |
| FWER p-Value | 0.743 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hao1 | 110 | 0.997 | 0.0427 | No |
| 2 | Cyp46a1 | 848 | 0.591 | 0.0394 | No |
| 3 | Dio1 | 1419 | 0.521 | 0.0399 | No |
| 4 | Cyp7b1 | 2413 | 0.398 | 0.0166 | No |
| 5 | Slc23a1 | 3000 | 0.361 | 0.0088 | No |
| 6 | Idi1 | 3052 | 0.357 | 0.0236 | No |
| 7 | Abca3 | 3584 | 0.308 | 0.0157 | No |
| 8 | Rbp1 | 3701 | 0.300 | 0.0250 | No |
| 9 | Prdx5 | 3960 | 0.282 | 0.0275 | No |
| 10 | Acsl1 | 4502 | 0.256 | 0.0166 | No |
| 11 | Akr1d1 | 4568 | 0.252 | 0.0258 | No |
| 12 | Mlycd | 4942 | 0.234 | 0.0211 | No |
| 13 | Abcd1 | 5612 | 0.209 | 0.0026 | No |
| 14 | Efhc1 | 5770 | 0.201 | 0.0055 | No |
| 15 | Nr3c2 | 6331 | 0.181 | -0.0098 | No |
| 16 | Abca2 | 6403 | 0.178 | -0.0043 | No |
| 17 | Lonp2 | 7244 | 0.145 | -0.0331 | No |
| 18 | Abcd2 | 7337 | 0.141 | -0.0304 | No |
| 19 | Acsl5 | 7424 | 0.138 | -0.0274 | No |
| 20 | Abca5 | 7572 | 0.133 | -0.0274 | No |
| 21 | Pex11g | 7794 | 0.125 | -0.0308 | No |
| 22 | Retsat | 7866 | 0.123 | -0.0280 | No |
| 23 | Pex16 | 8150 | 0.113 | -0.0347 | No |
| 24 | Slc27a5 | 8747 | 0.095 | -0.0555 | No |
| 25 | Bmp6 | 9040 | 0.086 | -0.0638 | No |
| 26 | Aqp9 | 9409 | 0.076 | -0.0758 | No |
| 27 | Pnpla8 | 9415 | 0.075 | -0.0725 | No |
| 28 | Dhcr24 | 9429 | 0.075 | -0.0695 | No |
| 29 | Slc29a1 | 9567 | 0.071 | -0.0719 | No |
| 30 | Hsd17b4 | 9674 | 0.068 | -0.0732 | No |
| 31 | Rxra | 9785 | 0.065 | -0.0748 | No |
| 32 | Pex6 | 9851 | 0.063 | -0.0746 | No |
| 33 | Lipe | 9853 | 0.063 | -0.0717 | No |
| 34 | Idh1 | 10134 | 0.055 | -0.0810 | No |
| 35 | Lck | 10205 | 0.052 | -0.0814 | No |
| 36 | Slc23a2 | 10577 | 0.042 | -0.0952 | No |
| 37 | Pex1 | 10810 | 0.036 | -0.1034 | No |
| 38 | Crot | 11761 | 0.008 | -0.1434 | No |
| 39 | Pex26 | 12371 | -0.002 | -0.1692 | No |
| 40 | Gclm | 12604 | -0.007 | -0.1787 | No |
| 41 | Gnmt | 12847 | -0.014 | -0.1883 | No |
| 42 | Phyh | 13065 | -0.020 | -0.1966 | No |
| 43 | Gc | 13120 | -0.021 | -0.1979 | No |
| 44 | Hsd17b11 | 13162 | -0.022 | -0.1986 | No |
| 45 | Pfkm | 13403 | -0.029 | -0.2074 | No |
| 46 | Pex7 | 13481 | -0.031 | -0.2092 | No |
| 47 | Scp2 | 13602 | -0.035 | -0.2127 | No |
| 48 | Idh2 | 13683 | -0.037 | -0.2143 | No |
| 49 | Dio2 | 14641 | -0.064 | -0.2519 | No |
| 50 | Nr1h4 | 14717 | -0.066 | -0.2520 | No |
| 51 | Optn | 15013 | -0.076 | -0.2609 | No |
| 52 | Tfcp2l1 | 15106 | -0.079 | -0.2610 | No |
| 53 | Aldh1a1 | 15257 | -0.085 | -0.2634 | No |
| 54 | Nedd4 | 15666 | -0.097 | -0.2761 | No |
| 55 | Pxmp2 | 15755 | -0.099 | -0.2752 | No |
| 56 | Amacr | 15764 | -0.099 | -0.2708 | No |
| 57 | Slc27a2 | 16521 | -0.125 | -0.2970 | No |
| 58 | Gnpat | 16524 | -0.125 | -0.2912 | No |
| 59 | Isoc1 | 16680 | -0.131 | -0.2916 | No |
| 60 | Hsd3b7 | 16889 | -0.138 | -0.2939 | No |
| 61 | Nudt12 | 17082 | -0.145 | -0.2952 | No |
| 62 | Abcd3 | 17214 | -0.150 | -0.2936 | No |
| 63 | Abca9 | 17603 | -0.164 | -0.3023 | Yes |
| 64 | Ar | 17676 | -0.167 | -0.2974 | Yes |
| 65 | Klf1 | 17803 | -0.171 | -0.2947 | Yes |
| 66 | Pex13 | 17909 | -0.176 | -0.2908 | Yes |
| 67 | Fdxr | 18093 | -0.182 | -0.2899 | Yes |
| 68 | Abca4 | 18128 | -0.182 | -0.2827 | Yes |
| 69 | Aldh9a1 | 18428 | -0.195 | -0.2862 | Yes |
| 70 | Atxn1 | 18557 | -0.200 | -0.2821 | Yes |
| 71 | Sod1 | 18830 | -0.210 | -0.2837 | Yes |
| 72 | Cyp27a1 | 19164 | -0.224 | -0.2872 | Yes |
| 73 | Fads1 | 19172 | -0.225 | -0.2768 | Yes |
| 74 | Abca8b | 19329 | -0.233 | -0.2724 | Yes |
| 75 | Abca6 | 19374 | -0.235 | -0.2631 | Yes |
| 76 | Abcg4 | 19384 | -0.235 | -0.2523 | Yes |
| 77 | Gstk1 | 19578 | -0.244 | -0.2489 | Yes |
| 78 | Fads2 | 19875 | -0.260 | -0.2492 | Yes |
| 79 | Bcar3 | 19954 | -0.264 | -0.2399 | Yes |
| 80 | Npc1 | 20010 | -0.267 | -0.2296 | Yes |
| 81 | Pex19 | 20443 | -0.291 | -0.2341 | Yes |
| 82 | Cat | 20608 | -0.301 | -0.2268 | Yes |
| 83 | Cyp39a1 | 21575 | -0.360 | -0.2508 | Yes |
| 84 | Soat2 | 21730 | -0.372 | -0.2397 | Yes |
| 85 | Ephx2 | 21808 | -0.379 | -0.2250 | Yes |
| 86 | Nr0b2 | 22340 | -0.433 | -0.2270 | Yes |
| 87 | Slc35b2 | 22477 | -0.451 | -0.2113 | Yes |
| 88 | Pipox | 22672 | -0.478 | -0.1969 | Yes |
| 89 | Hacl1 | 22744 | -0.490 | -0.1766 | Yes |
| 90 | Abca1 | 22949 | -0.527 | -0.1603 | Yes |
| 91 | Pex12 | 23037 | -0.542 | -0.1383 | Yes |
| 92 | Slc22a18 | 23110 | -0.559 | -0.1148 | Yes |
| 93 | Sult2b1 | 23339 | -0.630 | -0.0946 | Yes |
| 94 | Pex11a | 23394 | -0.667 | -0.0652 | Yes |
| 95 | Paox | 23530 | -0.778 | -0.0340 | Yes |
| 96 | Pecr | 23542 | -0.799 | 0.0035 | Yes |