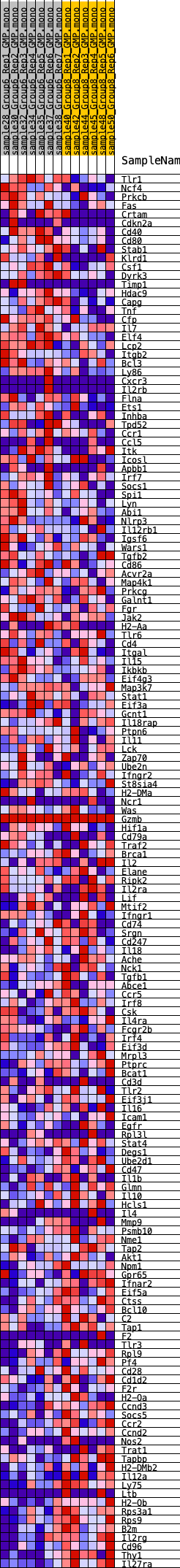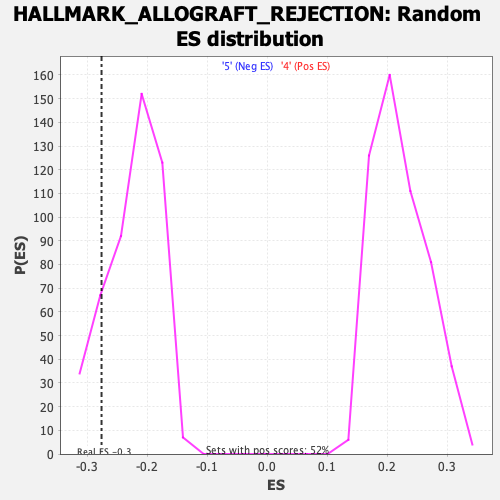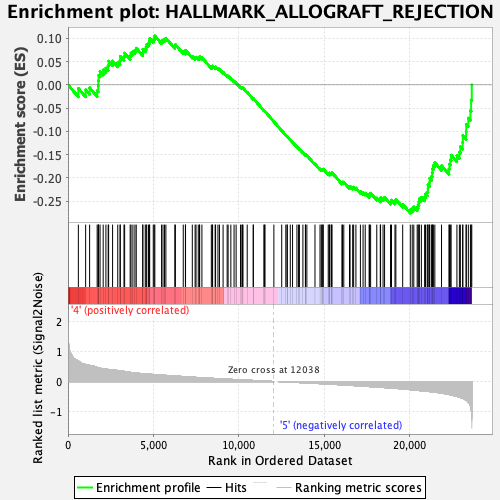
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group6_versus_Group8.GMP.mono_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | GMP.mono_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_ALLOGRAFT_REJECTION |
| Enrichment Score (ES) | -0.27580258 |
| Normalized Enrichment Score (NES) | -1.2329593 |
| Nominal p-value | 0.14526317 |
| FDR q-value | 0.57874787 |
| FWER p-Value | 0.876 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tlr1 | 606 | 0.679 | -0.0076 | No |
| 2 | Ncf4 | 1032 | 0.564 | -0.0107 | No |
| 3 | Prkcb | 1265 | 0.536 | -0.0062 | No |
| 4 | Fas | 1706 | 0.468 | -0.0124 | No |
| 5 | Crtam | 1770 | 0.456 | -0.0029 | No |
| 6 | Cdkn2a | 1776 | 0.454 | 0.0090 | No |
| 7 | Cd40 | 1792 | 0.451 | 0.0204 | No |
| 8 | Cd80 | 1875 | 0.440 | 0.0287 | No |
| 9 | Stab1 | 2058 | 0.420 | 0.0322 | No |
| 10 | Klrd1 | 2218 | 0.406 | 0.0363 | No |
| 11 | Csf1 | 2350 | 0.401 | 0.0415 | No |
| 12 | Dyrk3 | 2377 | 0.400 | 0.0510 | No |
| 13 | Timp1 | 2608 | 0.382 | 0.0515 | No |
| 14 | Hdac9 | 2911 | 0.363 | 0.0483 | No |
| 15 | Capg | 3046 | 0.358 | 0.0522 | No |
| 16 | Tnf | 3059 | 0.357 | 0.0612 | No |
| 17 | Cfp | 3275 | 0.336 | 0.0611 | No |
| 18 | Il7 | 3314 | 0.332 | 0.0683 | No |
| 19 | Elf4 | 3637 | 0.304 | 0.0628 | No |
| 20 | Lcp2 | 3687 | 0.301 | 0.0687 | No |
| 21 | Itgb2 | 3797 | 0.294 | 0.0720 | No |
| 22 | Bcl3 | 3915 | 0.286 | 0.0746 | No |
| 23 | Ly86 | 3993 | 0.281 | 0.0789 | No |
| 24 | Cxcr3 | 4373 | 0.261 | 0.0697 | No |
| 25 | Il2rb | 4377 | 0.261 | 0.0766 | No |
| 26 | Flna | 4518 | 0.255 | 0.0774 | No |
| 27 | Ets1 | 4571 | 0.252 | 0.0820 | No |
| 28 | Inhba | 4607 | 0.250 | 0.0872 | No |
| 29 | Tpd52 | 4711 | 0.248 | 0.0894 | No |
| 30 | Ccr1 | 4758 | 0.245 | 0.0940 | No |
| 31 | Ccl5 | 4778 | 0.244 | 0.0997 | No |
| 32 | Itk | 4988 | 0.231 | 0.0970 | No |
| 33 | Icosl | 5040 | 0.228 | 0.1009 | No |
| 34 | Apbb1 | 5080 | 0.226 | 0.1053 | No |
| 35 | Irf7 | 5473 | 0.216 | 0.0944 | No |
| 36 | Socs1 | 5558 | 0.212 | 0.0965 | No |
| 37 | Spi1 | 5647 | 0.207 | 0.0983 | No |
| 38 | Lyn | 5726 | 0.203 | 0.1004 | No |
| 39 | Abi1 | 6249 | 0.184 | 0.0831 | No |
| 40 | Nlrp3 | 6282 | 0.182 | 0.0866 | No |
| 41 | Il12rb1 | 6742 | 0.165 | 0.0715 | No |
| 42 | Igsf6 | 6869 | 0.160 | 0.0704 | No |
| 43 | Wars1 | 6876 | 0.160 | 0.0744 | No |
| 44 | Tgfb2 | 7272 | 0.144 | 0.0614 | No |
| 45 | Cd86 | 7436 | 0.138 | 0.0582 | No |
| 46 | Acvr2a | 7489 | 0.136 | 0.0596 | No |
| 47 | Map4k1 | 7621 | 0.131 | 0.0575 | No |
| 48 | Prkcg | 7694 | 0.128 | 0.0579 | No |
| 49 | Galnt1 | 7702 | 0.128 | 0.0610 | No |
| 50 | Fgr | 7834 | 0.124 | 0.0588 | No |
| 51 | Jak2 | 8385 | 0.107 | 0.0382 | No |
| 52 | H2-Aa | 8446 | 0.105 | 0.0384 | No |
| 53 | Tlr6 | 8460 | 0.104 | 0.0407 | No |
| 54 | Cd4 | 8608 | 0.099 | 0.0370 | No |
| 55 | Itgal | 8634 | 0.098 | 0.0386 | No |
| 56 | Il15 | 8773 | 0.094 | 0.0352 | No |
| 57 | Ikbkb | 8860 | 0.092 | 0.0340 | No |
| 58 | Eif4g3 | 9073 | 0.085 | 0.0273 | No |
| 59 | Map3k7 | 9311 | 0.079 | 0.0193 | No |
| 60 | Stat1 | 9367 | 0.077 | 0.0190 | No |
| 61 | Eif3a | 9527 | 0.072 | 0.0141 | No |
| 62 | Gcnt1 | 9715 | 0.067 | 0.0080 | No |
| 63 | Il18rap | 9835 | 0.063 | 0.0046 | No |
| 64 | Ptpn6 | 10091 | 0.056 | -0.0048 | No |
| 65 | Il11 | 10173 | 0.054 | -0.0068 | No |
| 66 | Lck | 10205 | 0.052 | -0.0067 | No |
| 67 | Zap70 | 10237 | 0.052 | -0.0067 | No |
| 68 | Ube2n | 10484 | 0.045 | -0.0160 | No |
| 69 | Ifngr2 | 10839 | 0.035 | -0.0301 | No |
| 70 | St8sia4 | 10840 | 0.035 | -0.0292 | No |
| 71 | H2-DMa | 11461 | 0.016 | -0.0552 | No |
| 72 | Ncr1 | 11490 | 0.015 | -0.0559 | No |
| 73 | Was | 11526 | 0.014 | -0.0571 | No |
| 74 | Gzmb | 12044 | 0.000 | -0.0791 | No |
| 75 | Hif1a | 12503 | -0.005 | -0.0985 | No |
| 76 | Cd79a | 12743 | -0.011 | -0.1084 | No |
| 77 | Traf2 | 12818 | -0.013 | -0.1112 | No |
| 78 | Brca1 | 12829 | -0.013 | -0.1112 | No |
| 79 | Il2 | 13004 | -0.018 | -0.1182 | No |
| 80 | Elane | 13137 | -0.022 | -0.1232 | No |
| 81 | Ripk2 | 13393 | -0.029 | -0.1333 | No |
| 82 | Il2ra | 13488 | -0.031 | -0.1365 | No |
| 83 | Lif | 13538 | -0.032 | -0.1377 | No |
| 84 | Mtif2 | 13742 | -0.039 | -0.1453 | No |
| 85 | Ifngr1 | 13890 | -0.043 | -0.1504 | No |
| 86 | Cd74 | 13892 | -0.043 | -0.1493 | No |
| 87 | Srgn | 13974 | -0.046 | -0.1515 | No |
| 88 | Cd247 | 14449 | -0.059 | -0.1701 | No |
| 89 | Il18 | 14744 | -0.067 | -0.1809 | No |
| 90 | Ache | 14820 | -0.070 | -0.1822 | No |
| 91 | Nck1 | 14869 | -0.072 | -0.1823 | No |
| 92 | Tgfb1 | 14908 | -0.073 | -0.1820 | No |
| 93 | Abce1 | 14933 | -0.074 | -0.1811 | No |
| 94 | Ccr5 | 15224 | -0.083 | -0.1912 | No |
| 95 | Irf8 | 15289 | -0.086 | -0.1916 | No |
| 96 | Csk | 15291 | -0.086 | -0.1894 | No |
| 97 | Il4ra | 15393 | -0.089 | -0.1913 | No |
| 98 | Fcgr2b | 15412 | -0.090 | -0.1896 | No |
| 99 | Irf4 | 15447 | -0.090 | -0.1887 | No |
| 100 | Eif3d | 16026 | -0.108 | -0.2104 | No |
| 101 | Mrpl3 | 16058 | -0.109 | -0.2088 | No |
| 102 | Ptprc | 16134 | -0.111 | -0.2090 | No |
| 103 | Bcat1 | 16469 | -0.123 | -0.2200 | No |
| 104 | Cd3d | 16501 | -0.124 | -0.2180 | No |
| 105 | Tlr2 | 16654 | -0.130 | -0.2210 | No |
| 106 | Eif3j1 | 16711 | -0.132 | -0.2198 | No |
| 107 | Il16 | 16844 | -0.136 | -0.2218 | No |
| 108 | Icam1 | 17106 | -0.146 | -0.2291 | No |
| 109 | Egfr | 17262 | -0.152 | -0.2316 | No |
| 110 | Rpl3l | 17388 | -0.157 | -0.2327 | No |
| 111 | Stat4 | 17618 | -0.165 | -0.2381 | No |
| 112 | Degs1 | 17636 | -0.165 | -0.2344 | No |
| 113 | Ube2d1 | 17704 | -0.168 | -0.2328 | No |
| 114 | Cd47 | 18064 | -0.181 | -0.2432 | No |
| 115 | Il1b | 18269 | -0.188 | -0.2469 | No |
| 116 | Glmn | 18286 | -0.189 | -0.2425 | No |
| 117 | Il10 | 18432 | -0.195 | -0.2435 | No |
| 118 | Hcls1 | 18520 | -0.199 | -0.2419 | No |
| 119 | Il4 | 18869 | -0.212 | -0.2510 | No |
| 120 | Mmp9 | 18925 | -0.215 | -0.2476 | No |
| 121 | Psmb10 | 19129 | -0.223 | -0.2503 | No |
| 122 | Nme1 | 19181 | -0.225 | -0.2464 | No |
| 123 | Tap2 | 19580 | -0.244 | -0.2569 | No |
| 124 | Akt1 | 20025 | -0.268 | -0.2686 | Yes |
| 125 | Npm1 | 20138 | -0.276 | -0.2660 | Yes |
| 126 | Gpr65 | 20230 | -0.281 | -0.2624 | Yes |
| 127 | Ifnar2 | 20428 | -0.290 | -0.2630 | Yes |
| 128 | Eif5a | 20485 | -0.293 | -0.2576 | Yes |
| 129 | Ctss | 20531 | -0.296 | -0.2516 | Yes |
| 130 | Bcl10 | 20554 | -0.297 | -0.2445 | Yes |
| 131 | C2 | 20673 | -0.304 | -0.2414 | Yes |
| 132 | Tap1 | 20864 | -0.317 | -0.2410 | Yes |
| 133 | F2 | 20913 | -0.319 | -0.2345 | Yes |
| 134 | Tlr3 | 21014 | -0.323 | -0.2301 | Yes |
| 135 | Rpl9 | 21058 | -0.326 | -0.2232 | Yes |
| 136 | Pf4 | 21062 | -0.327 | -0.2146 | Yes |
| 137 | Cd28 | 21156 | -0.332 | -0.2097 | Yes |
| 138 | Cd1d2 | 21159 | -0.332 | -0.2009 | Yes |
| 139 | F2r | 21273 | -0.341 | -0.1966 | Yes |
| 140 | H2-Oa | 21305 | -0.342 | -0.1888 | Yes |
| 141 | Ccnd3 | 21324 | -0.343 | -0.1803 | Yes |
| 142 | Socs5 | 21376 | -0.348 | -0.1732 | Yes |
| 143 | Ccr2 | 21460 | -0.352 | -0.1673 | Yes |
| 144 | Ccnd2 | 21852 | -0.384 | -0.1737 | Yes |
| 145 | Nos2 | 22288 | -0.428 | -0.1808 | Yes |
| 146 | Trat1 | 22315 | -0.431 | -0.1704 | Yes |
| 147 | Tapbp | 22368 | -0.437 | -0.1609 | Yes |
| 148 | H2-DMb2 | 22414 | -0.442 | -0.1510 | Yes |
| 149 | Il12a | 22753 | -0.491 | -0.1522 | Yes |
| 150 | Ly75 | 22897 | -0.516 | -0.1445 | Yes |
| 151 | Ltb | 22954 | -0.528 | -0.1328 | Yes |
| 152 | H2-Ob | 23089 | -0.553 | -0.1237 | Yes |
| 153 | Rps3a1 | 23098 | -0.555 | -0.1091 | Yes |
| 154 | Rps9 | 23292 | -0.617 | -0.1009 | Yes |
| 155 | B2m | 23299 | -0.618 | -0.0846 | Yes |
| 156 | Il2rg | 23421 | -0.682 | -0.0715 | Yes |
| 157 | Cd96 | 23543 | -0.802 | -0.0552 | Yes |
| 158 | Thy1 | 23584 | -0.914 | -0.0324 | Yes |
| 159 | Il27ra | 23622 | -1.273 | 0.0001 | Yes |