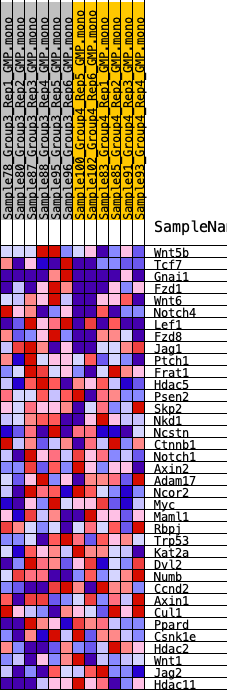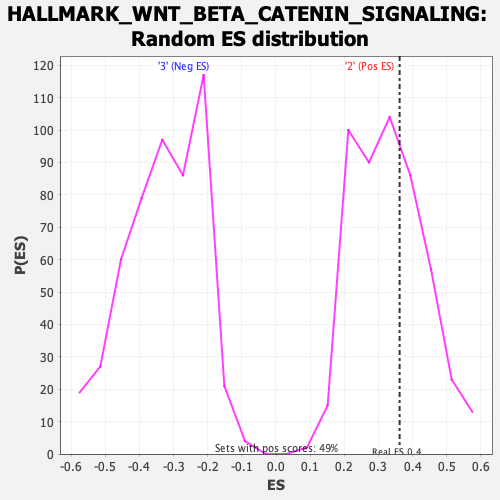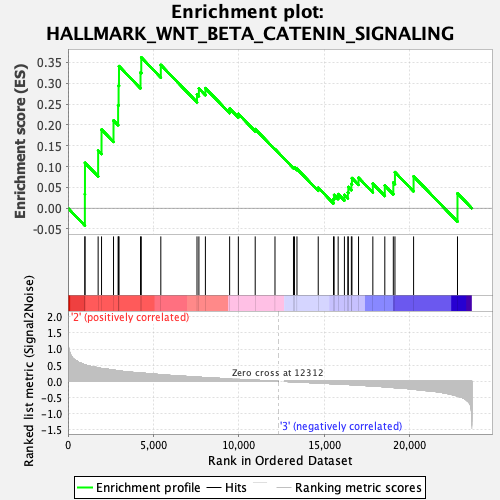
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group3_versus_Group4.GMP.mono_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_WNT_BETA_CATENIN_SIGNALING |
| Enrichment Score (ES) | 0.36211485 |
| Normalized Enrichment Score (NES) | 1.090854 |
| Nominal p-value | 0.37142858 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.97 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Wnt5b | 987 | 0.507 | 0.0338 | Yes |
| 2 | Tcf7 | 993 | 0.505 | 0.1089 | Yes |
| 3 | Gnai1 | 1763 | 0.418 | 0.1385 | Yes |
| 4 | Fzd1 | 1962 | 0.394 | 0.1889 | Yes |
| 5 | Wnt6 | 2663 | 0.346 | 0.2107 | Yes |
| 6 | Notch4 | 2932 | 0.324 | 0.2476 | Yes |
| 7 | Lef1 | 2960 | 0.322 | 0.2944 | Yes |
| 8 | Fzd8 | 2983 | 0.320 | 0.3412 | Yes |
| 9 | Jag1 | 4243 | 0.256 | 0.3260 | Yes |
| 10 | Ptch1 | 4286 | 0.254 | 0.3621 | Yes |
| 11 | Frat1 | 5430 | 0.204 | 0.3440 | No |
| 12 | Hdac5 | 7547 | 0.129 | 0.2735 | No |
| 13 | Psen2 | 7658 | 0.125 | 0.2875 | No |
| 14 | Skp2 | 8038 | 0.113 | 0.2883 | No |
| 15 | Nkd1 | 9456 | 0.074 | 0.2392 | No |
| 16 | Ncstn | 9966 | 0.060 | 0.2265 | No |
| 17 | Ctnnb1 | 10952 | 0.035 | 0.1899 | No |
| 18 | Notch1 | 12109 | 0.006 | 0.1418 | No |
| 19 | Axin2 | 13191 | -0.014 | 0.0980 | No |
| 20 | Adam17 | 13249 | -0.015 | 0.0978 | No |
| 21 | Ncor2 | 13395 | -0.019 | 0.0944 | No |
| 22 | Myc | 14639 | -0.050 | 0.0492 | No |
| 23 | Maml1 | 15532 | -0.073 | 0.0223 | No |
| 24 | Rbpj | 15569 | -0.074 | 0.0318 | No |
| 25 | Trp53 | 15805 | -0.080 | 0.0337 | No |
| 26 | Kat2a | 16171 | -0.091 | 0.0319 | No |
| 27 | Dvl2 | 16372 | -0.095 | 0.0377 | No |
| 28 | Numb | 16399 | -0.096 | 0.0509 | No |
| 29 | Ccnd2 | 16577 | -0.101 | 0.0585 | No |
| 30 | Axin1 | 16612 | -0.102 | 0.0723 | No |
| 31 | Cul1 | 16997 | -0.115 | 0.0731 | No |
| 32 | Ppard | 17834 | -0.142 | 0.0588 | No |
| 33 | Csnk1e | 18534 | -0.169 | 0.0544 | No |
| 34 | Hdac2 | 19032 | -0.190 | 0.0616 | No |
| 35 | Wnt1 | 19127 | -0.194 | 0.0866 | No |
| 36 | Jag2 | 20218 | -0.242 | 0.0765 | No |
| 37 | Hdac11 | 22786 | -0.456 | 0.0355 | No |