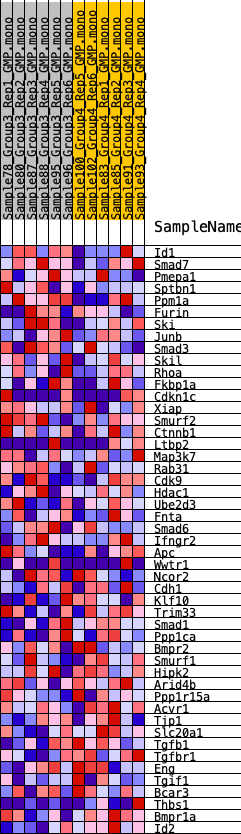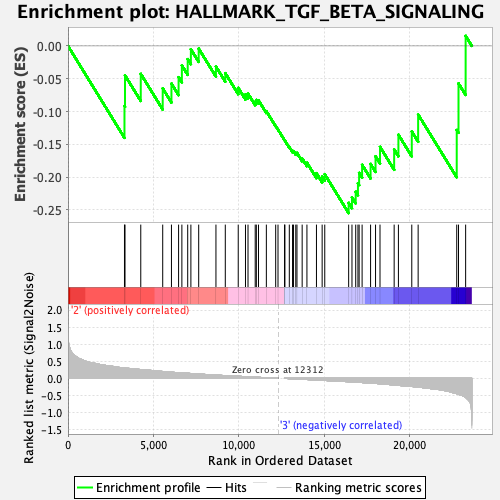
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group3_versus_Group4.GMP.mono_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_TGF_BETA_SIGNALING |
| Enrichment Score (ES) | -0.25483182 |
| Normalized Enrichment Score (NES) | -1.0793544 |
| Nominal p-value | 0.35852712 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.979 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Id1 | 3304 | 0.300 | -0.0916 | No |
| 2 | Smad7 | 3335 | 0.296 | -0.0449 | No |
| 3 | Pmepa1 | 4257 | 0.255 | -0.0426 | No |
| 4 | Sptbn1 | 5541 | 0.199 | -0.0647 | No |
| 5 | Ppm1a | 6052 | 0.179 | -0.0574 | No |
| 6 | Furin | 6470 | 0.169 | -0.0477 | No |
| 7 | Ski | 6662 | 0.161 | -0.0298 | No |
| 8 | Junb | 7003 | 0.149 | -0.0201 | No |
| 9 | Smad3 | 7189 | 0.141 | -0.0051 | No |
| 10 | Skil | 7646 | 0.126 | -0.0041 | No |
| 11 | Rhoa | 8655 | 0.096 | -0.0313 | No |
| 12 | Fkbp1a | 9202 | 0.080 | -0.0415 | No |
| 13 | Cdkn1c | 9959 | 0.060 | -0.0638 | No |
| 14 | Xiap | 10383 | 0.049 | -0.0738 | No |
| 15 | Smurf2 | 10531 | 0.046 | -0.0726 | No |
| 16 | Ctnnb1 | 10952 | 0.035 | -0.0848 | No |
| 17 | Ltbp2 | 11018 | 0.033 | -0.0823 | No |
| 18 | Map3k7 | 11144 | 0.029 | -0.0829 | No |
| 19 | Rab31 | 11604 | 0.018 | -0.0994 | No |
| 20 | Cdk9 | 12150 | 0.004 | -0.1219 | No |
| 21 | Hdac1 | 12286 | 0.001 | -0.1275 | No |
| 22 | Ube2d3 | 12668 | -0.000 | -0.1436 | No |
| 23 | Fnta | 12689 | -0.001 | -0.1444 | No |
| 24 | Smad6 | 12947 | -0.008 | -0.1541 | No |
| 25 | Ifngr2 | 13136 | -0.012 | -0.1601 | No |
| 26 | Apc | 13194 | -0.014 | -0.1603 | No |
| 27 | Wwtr1 | 13326 | -0.017 | -0.1631 | No |
| 28 | Ncor2 | 13395 | -0.019 | -0.1630 | No |
| 29 | Cdh1 | 13699 | -0.026 | -0.1716 | No |
| 30 | Klf10 | 13978 | -0.033 | -0.1780 | No |
| 31 | Trim33 | 14535 | -0.047 | -0.1939 | No |
| 32 | Smad1 | 14868 | -0.056 | -0.1989 | No |
| 33 | Ppp1ca | 15021 | -0.060 | -0.1956 | No |
| 34 | Bmpr2 | 16419 | -0.097 | -0.2392 | Yes |
| 35 | Smurf1 | 16613 | -0.102 | -0.2308 | Yes |
| 36 | Hipk2 | 16832 | -0.110 | -0.2222 | Yes |
| 37 | Arid4b | 16963 | -0.114 | -0.2094 | Yes |
| 38 | Ppp1r15a | 17032 | -0.116 | -0.1935 | Yes |
| 39 | Acvr1 | 17206 | -0.122 | -0.1811 | Yes |
| 40 | Tjp1 | 17702 | -0.137 | -0.1798 | Yes |
| 41 | Slc20a1 | 17992 | -0.148 | -0.1682 | Yes |
| 42 | Tgfb1 | 18253 | -0.157 | -0.1538 | Yes |
| 43 | Tgfbr1 | 19081 | -0.193 | -0.1577 | Yes |
| 44 | Eng | 19330 | -0.203 | -0.1353 | Yes |
| 45 | Tgif1 | 20112 | -0.235 | -0.1304 | Yes |
| 46 | Bcar3 | 20484 | -0.257 | -0.1046 | Yes |
| 47 | Thbs1 | 22742 | -0.446 | -0.1280 | Yes |
| 48 | Bmpr1a | 22846 | -0.465 | -0.0571 | Yes |
| 49 | Id2 | 23263 | -0.556 | 0.0153 | Yes |