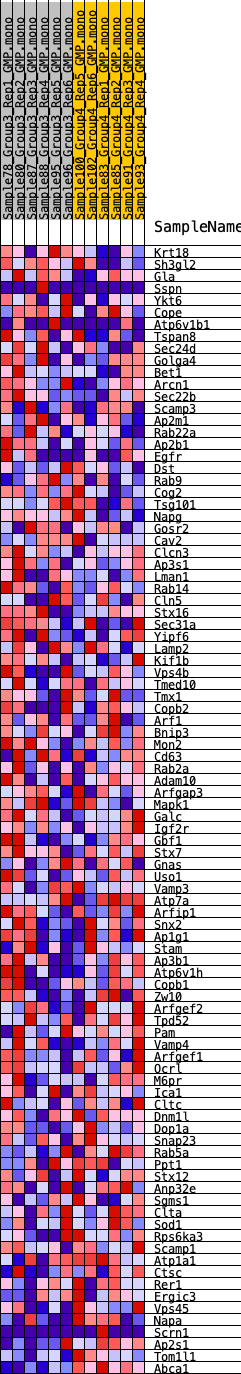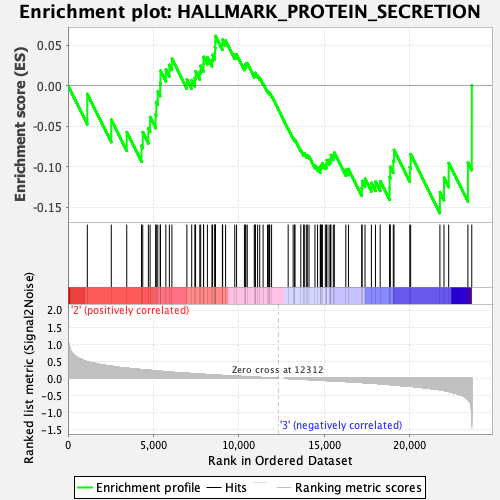
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group3_versus_Group4.GMP.mono_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | -0.15735336 |
| Normalized Enrichment Score (NES) | -0.77125883 |
| Nominal p-value | 0.6986028 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Krt18 | 1130 | 0.483 | -0.0102 | No |
| 2 | Sh3gl2 | 2532 | 0.357 | -0.0419 | No |
| 3 | Gla | 3437 | 0.293 | -0.0574 | No |
| 4 | Sspn | 4299 | 0.254 | -0.0741 | No |
| 5 | Ykt6 | 4373 | 0.253 | -0.0574 | No |
| 6 | Cope | 4702 | 0.238 | -0.0528 | No |
| 7 | Atp6v1b1 | 4802 | 0.234 | -0.0387 | No |
| 8 | Tspan8 | 5128 | 0.218 | -0.0355 | No |
| 9 | Sec24d | 5165 | 0.216 | -0.0201 | No |
| 10 | Golga4 | 5253 | 0.212 | -0.0072 | No |
| 11 | Bet1 | 5393 | 0.205 | 0.0030 | No |
| 12 | Arcn1 | 5410 | 0.204 | 0.0183 | No |
| 13 | Sec22b | 5725 | 0.191 | 0.0199 | No |
| 14 | Scamp3 | 5930 | 0.184 | 0.0256 | No |
| 15 | Ap2m1 | 6079 | 0.177 | 0.0332 | No |
| 16 | Rab22a | 6956 | 0.150 | 0.0077 | No |
| 17 | Ap2b1 | 7237 | 0.139 | 0.0067 | No |
| 18 | Egfr | 7422 | 0.133 | 0.0093 | No |
| 19 | Dst | 7463 | 0.132 | 0.0178 | No |
| 20 | Rab9 | 7711 | 0.124 | 0.0170 | No |
| 21 | Cog2 | 7763 | 0.122 | 0.0244 | No |
| 22 | Tsg101 | 7930 | 0.117 | 0.0265 | No |
| 23 | Napg | 7933 | 0.116 | 0.0355 | No |
| 24 | Gosr2 | 8155 | 0.110 | 0.0347 | No |
| 25 | Cav2 | 8422 | 0.103 | 0.0314 | No |
| 26 | Clcn3 | 8459 | 0.102 | 0.0379 | No |
| 27 | Ap3s1 | 8592 | 0.098 | 0.0399 | No |
| 28 | Lman1 | 8599 | 0.098 | 0.0473 | No |
| 29 | Rab14 | 8619 | 0.097 | 0.0541 | No |
| 30 | Cln5 | 8627 | 0.096 | 0.0613 | No |
| 31 | Stx16 | 9032 | 0.084 | 0.0508 | No |
| 32 | Sec31a | 9047 | 0.084 | 0.0567 | No |
| 33 | Yipf6 | 9220 | 0.080 | 0.0557 | No |
| 34 | Lamp2 | 9755 | 0.066 | 0.0382 | No |
| 35 | Kif1b | 9861 | 0.063 | 0.0386 | No |
| 36 | Vps4b | 10321 | 0.051 | 0.0231 | No |
| 37 | Tmed10 | 10370 | 0.049 | 0.0249 | No |
| 38 | Tmx1 | 10406 | 0.049 | 0.0272 | No |
| 39 | Copb2 | 10486 | 0.046 | 0.0275 | No |
| 40 | Arf1 | 10897 | 0.036 | 0.0129 | No |
| 41 | Bnip3 | 10951 | 0.035 | 0.0134 | No |
| 42 | Mon2 | 10961 | 0.034 | 0.0157 | No |
| 43 | Cd63 | 11092 | 0.031 | 0.0125 | No |
| 44 | Rab2a | 11217 | 0.028 | 0.0094 | No |
| 45 | Adam10 | 11414 | 0.023 | 0.0029 | No |
| 46 | Arfgap3 | 11672 | 0.016 | -0.0068 | No |
| 47 | Mapk1 | 11738 | 0.015 | -0.0084 | No |
| 48 | Galc | 11790 | 0.013 | -0.0096 | No |
| 49 | Igf2r | 11801 | 0.013 | -0.0090 | No |
| 50 | Gbf1 | 11910 | 0.010 | -0.0128 | No |
| 51 | Stx7 | 12880 | -0.006 | -0.0535 | No |
| 52 | Gnas | 13172 | -0.013 | -0.0649 | No |
| 53 | Uso1 | 13262 | -0.015 | -0.0675 | No |
| 54 | Vamp3 | 13276 | -0.016 | -0.0668 | No |
| 55 | Atp7a | 13620 | -0.025 | -0.0794 | No |
| 56 | Arfip1 | 13787 | -0.029 | -0.0842 | No |
| 57 | Snx2 | 13823 | -0.030 | -0.0834 | No |
| 58 | Ap1g1 | 13941 | -0.032 | -0.0858 | No |
| 59 | Stam | 14001 | -0.034 | -0.0857 | No |
| 60 | Ap3b1 | 14101 | -0.036 | -0.0871 | No |
| 61 | Atp6v1h | 14448 | -0.045 | -0.0983 | No |
| 62 | Copb1 | 14600 | -0.049 | -0.1009 | No |
| 63 | Zw10 | 14758 | -0.053 | -0.1034 | No |
| 64 | Arfgef2 | 14772 | -0.053 | -0.0998 | No |
| 65 | Tpd52 | 14831 | -0.055 | -0.0979 | No |
| 66 | Pam | 14882 | -0.057 | -0.0956 | No |
| 67 | Vamp4 | 15054 | -0.061 | -0.0981 | No |
| 68 | Arfgef1 | 15129 | -0.064 | -0.0963 | No |
| 69 | Ocrl | 15142 | -0.064 | -0.0918 | No |
| 70 | M6pr | 15275 | -0.067 | -0.0921 | No |
| 71 | Ica1 | 15359 | -0.069 | -0.0903 | No |
| 72 | Cltc | 15378 | -0.069 | -0.0856 | No |
| 73 | Dnm1l | 15519 | -0.073 | -0.0859 | No |
| 74 | Dop1a | 15581 | -0.074 | -0.0827 | No |
| 75 | Snap23 | 16253 | -0.093 | -0.1039 | No |
| 76 | Rab5a | 16407 | -0.096 | -0.1028 | No |
| 77 | Ppt1 | 17188 | -0.121 | -0.1265 | No |
| 78 | Stx12 | 17205 | -0.122 | -0.1176 | No |
| 79 | Anp32e | 17376 | -0.128 | -0.1149 | No |
| 80 | Sgms1 | 17750 | -0.139 | -0.1199 | No |
| 81 | Clta | 17988 | -0.147 | -0.1184 | No |
| 82 | Sod1 | 18264 | -0.158 | -0.1178 | No |
| 83 | Rps6ka3 | 18814 | -0.181 | -0.1269 | No |
| 84 | Scamp1 | 18817 | -0.182 | -0.1128 | No |
| 85 | Atp1a1 | 18855 | -0.183 | -0.1001 | No |
| 86 | Ctsc | 19031 | -0.190 | -0.0926 | No |
| 87 | Rer1 | 19072 | -0.192 | -0.0793 | No |
| 88 | Ergic3 | 19996 | -0.229 | -0.1007 | No |
| 89 | Vps45 | 20045 | -0.231 | -0.0846 | No |
| 90 | Napa | 21758 | -0.333 | -0.1313 | Yes |
| 91 | Scrn1 | 21997 | -0.354 | -0.1137 | Yes |
| 92 | Ap2s1 | 22270 | -0.382 | -0.0954 | Yes |
| 93 | Tom1l1 | 23395 | -0.619 | -0.0948 | Yes |
| 94 | Abca1 | 23621 | -1.335 | 0.0001 | Yes |