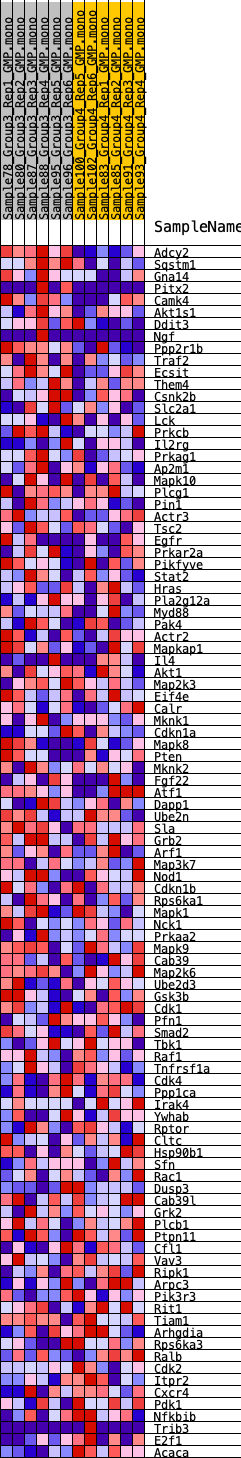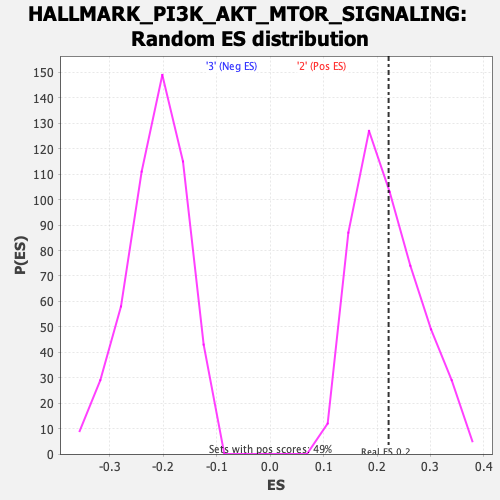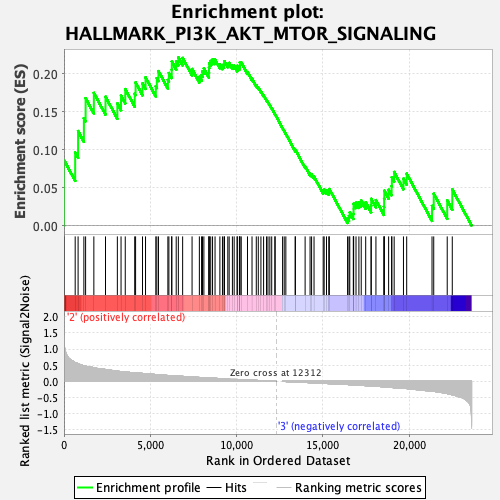
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group3_versus_Group4.GMP.mono_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | 0.2215882 |
| Normalized Enrichment Score (NES) | 1.013518 |
| Nominal p-value | 0.42798355 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.988 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Adcy2 | 3 | 1.333 | 0.0862 | Yes |
| 2 | Sqstm1 | 648 | 0.578 | 0.0963 | Yes |
| 3 | Gna14 | 817 | 0.546 | 0.1245 | Yes |
| 4 | Pitx2 | 1152 | 0.480 | 0.1414 | Yes |
| 5 | Camk4 | 1252 | 0.473 | 0.1678 | Yes |
| 6 | Akt1s1 | 1730 | 0.421 | 0.1749 | Yes |
| 7 | Ddit3 | 2405 | 0.365 | 0.1698 | Yes |
| 8 | Ngf | 3092 | 0.313 | 0.1610 | Yes |
| 9 | Ppp2r1b | 3306 | 0.300 | 0.1713 | Yes |
| 10 | Traf2 | 3550 | 0.291 | 0.1798 | Yes |
| 11 | Ecsit | 4098 | 0.266 | 0.1738 | Yes |
| 12 | Them4 | 4149 | 0.262 | 0.1886 | Yes |
| 13 | Csnk2b | 4550 | 0.243 | 0.1874 | Yes |
| 14 | Slc2a1 | 4725 | 0.236 | 0.1953 | Yes |
| 15 | Lck | 5321 | 0.209 | 0.1835 | Yes |
| 16 | Prkcb | 5381 | 0.206 | 0.1943 | Yes |
| 17 | Il2rg | 5475 | 0.203 | 0.2035 | Yes |
| 18 | Prkag1 | 6020 | 0.180 | 0.1921 | Yes |
| 19 | Ap2m1 | 6079 | 0.177 | 0.2011 | Yes |
| 20 | Mapk10 | 6241 | 0.172 | 0.2054 | Yes |
| 21 | Plcg1 | 6248 | 0.171 | 0.2162 | Yes |
| 22 | Pin1 | 6503 | 0.168 | 0.2163 | Yes |
| 23 | Actr3 | 6627 | 0.162 | 0.2216 | Yes |
| 24 | Tsc2 | 6876 | 0.153 | 0.2209 | No |
| 25 | Egfr | 7422 | 0.133 | 0.2064 | No |
| 26 | Prkar2a | 7845 | 0.119 | 0.1962 | No |
| 27 | Pikfyve | 7972 | 0.115 | 0.1983 | No |
| 28 | Stat2 | 8027 | 0.113 | 0.2033 | No |
| 29 | Hras | 8107 | 0.111 | 0.2071 | No |
| 30 | Pla2g12a | 8394 | 0.104 | 0.2017 | No |
| 31 | Myd88 | 8404 | 0.104 | 0.2080 | No |
| 32 | Pak4 | 8417 | 0.103 | 0.2142 | No |
| 33 | Actr2 | 8504 | 0.101 | 0.2171 | No |
| 34 | Mapkap1 | 8609 | 0.097 | 0.2190 | No |
| 35 | Il4 | 8754 | 0.093 | 0.2189 | No |
| 36 | Akt1 | 9034 | 0.084 | 0.2125 | No |
| 37 | Map2k3 | 9184 | 0.081 | 0.2114 | No |
| 38 | Eif4e | 9273 | 0.078 | 0.2127 | No |
| 39 | Calr | 9302 | 0.078 | 0.2165 | No |
| 40 | Mknk1 | 9494 | 0.073 | 0.2131 | No |
| 41 | Cdkn1a | 9572 | 0.071 | 0.2145 | No |
| 42 | Mapk8 | 9760 | 0.066 | 0.2108 | No |
| 43 | Pten | 9862 | 0.063 | 0.2106 | No |
| 44 | Mknk2 | 10036 | 0.058 | 0.2070 | No |
| 45 | Fgf22 | 10041 | 0.058 | 0.2106 | No |
| 46 | Atf1 | 10166 | 0.055 | 0.2089 | No |
| 47 | Dapp1 | 10191 | 0.055 | 0.2114 | No |
| 48 | Ube2n | 10192 | 0.055 | 0.2150 | No |
| 49 | Sla | 10272 | 0.052 | 0.2150 | No |
| 50 | Grb2 | 10634 | 0.043 | 0.2024 | No |
| 51 | Arf1 | 10897 | 0.036 | 0.1937 | No |
| 52 | Map3k7 | 11144 | 0.029 | 0.1851 | No |
| 53 | Nod1 | 11260 | 0.027 | 0.1819 | No |
| 54 | Cdkn1b | 11413 | 0.023 | 0.1769 | No |
| 55 | Rps6ka1 | 11568 | 0.019 | 0.1716 | No |
| 56 | Mapk1 | 11738 | 0.015 | 0.1654 | No |
| 57 | Nck1 | 11821 | 0.012 | 0.1627 | No |
| 58 | Prkaa2 | 11930 | 0.010 | 0.1587 | No |
| 59 | Mapk9 | 12038 | 0.007 | 0.1546 | No |
| 60 | Cab39 | 12221 | 0.002 | 0.1470 | No |
| 61 | Map2k6 | 12237 | 0.002 | 0.1465 | No |
| 62 | Ube2d3 | 12668 | -0.000 | 0.1282 | No |
| 63 | Gsk3b | 12762 | -0.003 | 0.1245 | No |
| 64 | Cdk1 | 12857 | -0.005 | 0.1208 | No |
| 65 | Pfn1 | 13398 | -0.019 | 0.0991 | No |
| 66 | Smad2 | 13410 | -0.019 | 0.0998 | No |
| 67 | Tbk1 | 13974 | -0.033 | 0.0780 | No |
| 68 | Raf1 | 14261 | -0.040 | 0.0685 | No |
| 69 | Tnfrsf1a | 14349 | -0.042 | 0.0675 | No |
| 70 | Cdk4 | 14491 | -0.046 | 0.0645 | No |
| 71 | Ppp1ca | 15021 | -0.060 | 0.0459 | No |
| 72 | Irak4 | 15077 | -0.062 | 0.0476 | No |
| 73 | Ywhab | 15213 | -0.066 | 0.0461 | No |
| 74 | Rptor | 15347 | -0.068 | 0.0449 | No |
| 75 | Cltc | 15378 | -0.069 | 0.0481 | No |
| 76 | Hsp90b1 | 16434 | -0.097 | 0.0096 | No |
| 77 | Sfn | 16515 | -0.100 | 0.0126 | No |
| 78 | Rac1 | 16562 | -0.101 | 0.0172 | No |
| 79 | Dusp3 | 16767 | -0.107 | 0.0155 | No |
| 80 | Cab39l | 16783 | -0.108 | 0.0218 | No |
| 81 | Grk2 | 16784 | -0.108 | 0.0288 | No |
| 82 | Plcb1 | 16924 | -0.113 | 0.0302 | No |
| 83 | Ptpn11 | 17091 | -0.118 | 0.0308 | No |
| 84 | Cfl1 | 17226 | -0.122 | 0.0330 | No |
| 85 | Vav3 | 17485 | -0.131 | 0.0306 | No |
| 86 | Ripk1 | 17797 | -0.141 | 0.0265 | No |
| 87 | Arpc3 | 17805 | -0.141 | 0.0353 | No |
| 88 | Pik3r3 | 18078 | -0.151 | 0.0335 | No |
| 89 | Rit1 | 18541 | -0.169 | 0.0248 | No |
| 90 | Tiam1 | 18554 | -0.170 | 0.0353 | No |
| 91 | Arhgdia | 18564 | -0.170 | 0.0459 | No |
| 92 | Rps6ka3 | 18814 | -0.181 | 0.0471 | No |
| 93 | Ralb | 18987 | -0.188 | 0.0520 | No |
| 94 | Cdk2 | 19002 | -0.188 | 0.0636 | No |
| 95 | Itpr2 | 19135 | -0.195 | 0.0706 | No |
| 96 | Cxcr4 | 19674 | -0.218 | 0.0618 | No |
| 97 | Pdk1 | 19861 | -0.222 | 0.0683 | No |
| 98 | Nfkbib | 21329 | -0.308 | 0.0259 | No |
| 99 | Trib3 | 21428 | -0.313 | 0.0420 | No |
| 100 | E2f1 | 22210 | -0.377 | 0.0332 | No |
| 101 | Acaca | 22503 | -0.415 | 0.0477 | No |