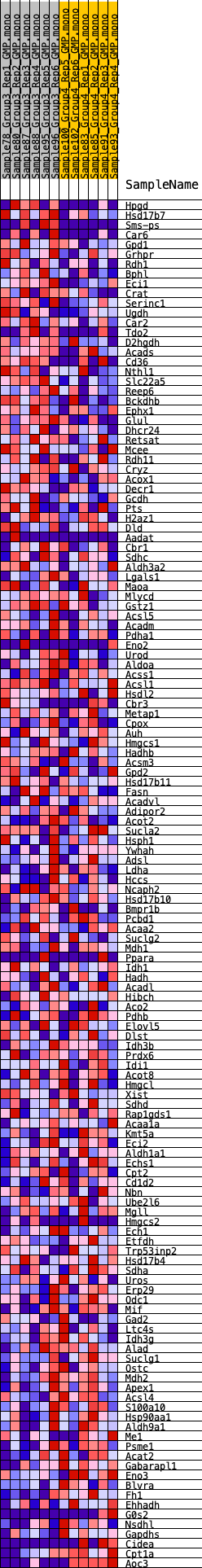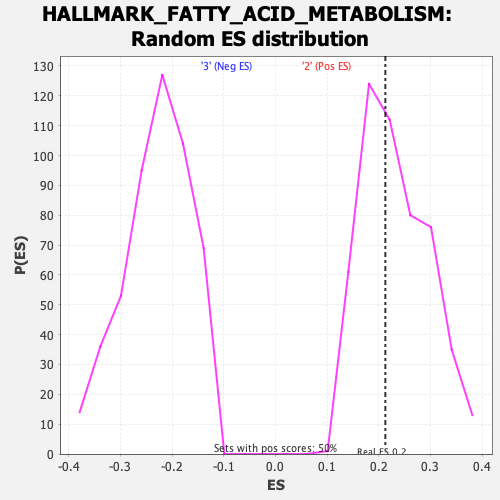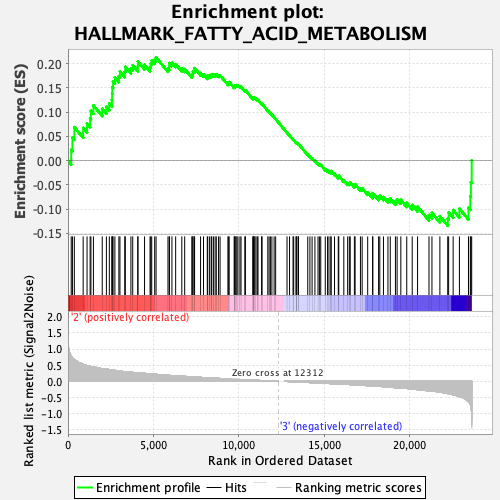
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group3_versus_Group4.GMP.mono_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_FATTY_ACID_METABOLISM |
| Enrichment Score (ES) | 0.21281604 |
| Normalized Enrichment Score (NES) | 0.91271466 |
| Nominal p-value | 0.57768923 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hpgd | 188 | 0.788 | 0.0225 | Yes |
| 2 | Hsd17b7 | 259 | 0.731 | 0.0477 | Yes |
| 3 | Sms-ps | 370 | 0.672 | 0.0690 | Yes |
| 4 | Car6 | 887 | 0.530 | 0.0675 | Yes |
| 5 | Gpd1 | 1114 | 0.485 | 0.0767 | Yes |
| 6 | Grhpr | 1294 | 0.463 | 0.0870 | Yes |
| 7 | Rdh1 | 1340 | 0.458 | 0.1027 | Yes |
| 8 | Bphl | 1487 | 0.447 | 0.1138 | Yes |
| 9 | Eci1 | 2003 | 0.391 | 0.1070 | Yes |
| 10 | Crat | 2246 | 0.374 | 0.1111 | Yes |
| 11 | Serinc1 | 2417 | 0.364 | 0.1180 | Yes |
| 12 | Ugdh | 2569 | 0.354 | 0.1252 | Yes |
| 13 | Car2 | 2584 | 0.353 | 0.1383 | Yes |
| 14 | Tdo2 | 2594 | 0.352 | 0.1515 | Yes |
| 15 | D2hgdh | 2637 | 0.348 | 0.1631 | Yes |
| 16 | Acads | 2743 | 0.340 | 0.1718 | Yes |
| 17 | Cd36 | 2977 | 0.320 | 0.1743 | Yes |
| 18 | Nthl1 | 3039 | 0.316 | 0.1839 | Yes |
| 19 | Slc22a5 | 3321 | 0.297 | 0.1834 | Yes |
| 20 | Reep6 | 3352 | 0.295 | 0.1936 | Yes |
| 21 | Bckdhb | 3681 | 0.282 | 0.1905 | Yes |
| 22 | Ephx1 | 3787 | 0.276 | 0.1967 | Yes |
| 23 | Glul | 4086 | 0.266 | 0.1943 | Yes |
| 24 | Dhcr24 | 4090 | 0.266 | 0.2044 | Yes |
| 25 | Retsat | 4478 | 0.247 | 0.1975 | Yes |
| 26 | Mcee | 4800 | 0.234 | 0.1929 | Yes |
| 27 | Rdh11 | 4841 | 0.232 | 0.2001 | Yes |
| 28 | Cryz | 4898 | 0.229 | 0.2066 | Yes |
| 29 | Acox1 | 5072 | 0.220 | 0.2078 | Yes |
| 30 | Decr1 | 5151 | 0.217 | 0.2128 | Yes |
| 31 | Gcdh | 5849 | 0.187 | 0.1903 | No |
| 32 | Pts | 5929 | 0.184 | 0.1941 | No |
| 33 | H2az1 | 5931 | 0.184 | 0.2012 | No |
| 34 | Dld | 6080 | 0.177 | 0.2017 | No |
| 35 | Aadat | 6298 | 0.171 | 0.1991 | No |
| 36 | Cbr1 | 6658 | 0.161 | 0.1900 | No |
| 37 | Sdhc | 6827 | 0.154 | 0.1888 | No |
| 38 | Aldh3a2 | 7243 | 0.139 | 0.1765 | No |
| 39 | Lgals1 | 7299 | 0.137 | 0.1795 | No |
| 40 | Maoa | 7311 | 0.137 | 0.1843 | No |
| 41 | Mlycd | 7390 | 0.134 | 0.1862 | No |
| 42 | Gstz1 | 7406 | 0.133 | 0.1907 | No |
| 43 | Acsl5 | 7756 | 0.122 | 0.1805 | No |
| 44 | Acadm | 7928 | 0.117 | 0.1778 | No |
| 45 | Pdha1 | 8149 | 0.110 | 0.1726 | No |
| 46 | Eno2 | 8174 | 0.109 | 0.1758 | No |
| 47 | Urod | 8284 | 0.108 | 0.1753 | No |
| 48 | Aldoa | 8379 | 0.105 | 0.1754 | No |
| 49 | Acss1 | 8409 | 0.104 | 0.1781 | No |
| 50 | Acsl1 | 8507 | 0.100 | 0.1779 | No |
| 51 | Hsdl2 | 8605 | 0.097 | 0.1775 | No |
| 52 | Cbr3 | 8676 | 0.095 | 0.1782 | No |
| 53 | Metap1 | 8808 | 0.091 | 0.1762 | No |
| 54 | Cpox | 8896 | 0.088 | 0.1759 | No |
| 55 | Auh | 9368 | 0.076 | 0.1588 | No |
| 56 | Hmgcs1 | 9369 | 0.076 | 0.1617 | No |
| 57 | Hadhb | 9429 | 0.074 | 0.1620 | No |
| 58 | Acsm3 | 9720 | 0.067 | 0.1523 | No |
| 59 | Gpd2 | 9757 | 0.066 | 0.1533 | No |
| 60 | Hsd17b11 | 9767 | 0.066 | 0.1555 | No |
| 61 | Fasn | 9821 | 0.064 | 0.1557 | No |
| 62 | Acadvl | 9879 | 0.063 | 0.1557 | No |
| 63 | Adipor2 | 9943 | 0.061 | 0.1553 | No |
| 64 | Acot2 | 10058 | 0.058 | 0.1527 | No |
| 65 | Sucla2 | 10122 | 0.056 | 0.1522 | No |
| 66 | Hsph1 | 10347 | 0.050 | 0.1446 | No |
| 67 | Ywhah | 10379 | 0.049 | 0.1452 | No |
| 68 | Adsl | 10807 | 0.039 | 0.1285 | No |
| 69 | Ldha | 10848 | 0.038 | 0.1283 | No |
| 70 | Hccs | 10858 | 0.037 | 0.1293 | No |
| 71 | Ncaph2 | 10867 | 0.037 | 0.1304 | No |
| 72 | Hsd17b10 | 10922 | 0.036 | 0.1295 | No |
| 73 | Bmpr1b | 10995 | 0.033 | 0.1277 | No |
| 74 | Pcbd1 | 11072 | 0.031 | 0.1257 | No |
| 75 | Acaa2 | 11116 | 0.030 | 0.1250 | No |
| 76 | Suclg2 | 11324 | 0.025 | 0.1172 | No |
| 77 | Mdh1 | 11353 | 0.024 | 0.1169 | No |
| 78 | Ppara | 11689 | 0.016 | 0.1033 | No |
| 79 | Idh1 | 11765 | 0.014 | 0.1006 | No |
| 80 | Hadh | 11841 | 0.012 | 0.0979 | No |
| 81 | Acadl | 11928 | 0.010 | 0.0946 | No |
| 82 | Hibch | 12058 | 0.007 | 0.0894 | No |
| 83 | Aco2 | 12148 | 0.004 | 0.0857 | No |
| 84 | Pdhb | 12813 | -0.004 | 0.0576 | No |
| 85 | Elovl5 | 12961 | -0.008 | 0.0517 | No |
| 86 | Dlst | 13165 | -0.013 | 0.0435 | No |
| 87 | Idh3b | 13205 | -0.014 | 0.0424 | No |
| 88 | Prdx6 | 13348 | -0.017 | 0.0370 | No |
| 89 | Idi1 | 13349 | -0.017 | 0.0377 | No |
| 90 | Acot8 | 13393 | -0.019 | 0.0366 | No |
| 91 | Hmgcl | 13480 | -0.021 | 0.0337 | No |
| 92 | Xist | 13481 | -0.021 | 0.0345 | No |
| 93 | Sdhd | 14027 | -0.035 | 0.0127 | No |
| 94 | Rap1gds1 | 14153 | -0.037 | 0.0088 | No |
| 95 | Acaa1a | 14280 | -0.041 | 0.0050 | No |
| 96 | Kmt5a | 14445 | -0.045 | -0.0003 | No |
| 97 | Eci2 | 14627 | -0.050 | -0.0061 | No |
| 98 | Aldh1a1 | 14708 | -0.052 | -0.0074 | No |
| 99 | Echs1 | 14785 | -0.054 | -0.0086 | No |
| 100 | Cpt2 | 15051 | -0.061 | -0.0175 | No |
| 101 | Cd1d2 | 15184 | -0.065 | -0.0206 | No |
| 102 | Nbn | 15267 | -0.067 | -0.0215 | No |
| 103 | Ube2l6 | 15379 | -0.069 | -0.0236 | No |
| 104 | Mgll | 15392 | -0.070 | -0.0214 | No |
| 105 | Hmgcs2 | 15593 | -0.075 | -0.0270 | No |
| 106 | Ech1 | 15811 | -0.080 | -0.0332 | No |
| 107 | Etfdh | 15841 | -0.081 | -0.0313 | No |
| 108 | Trp53inp2 | 16136 | -0.090 | -0.0403 | No |
| 109 | Hsd17b4 | 16361 | -0.095 | -0.0462 | No |
| 110 | Sdha | 16458 | -0.098 | -0.0465 | No |
| 111 | Uros | 16511 | -0.100 | -0.0448 | No |
| 112 | Erp29 | 16754 | -0.107 | -0.0510 | No |
| 113 | Odc1 | 16796 | -0.108 | -0.0485 | No |
| 114 | Mif | 17108 | -0.118 | -0.0572 | No |
| 115 | Gad2 | 17209 | -0.122 | -0.0568 | No |
| 116 | Ltc4s | 17533 | -0.133 | -0.0654 | No |
| 117 | Idh3g | 17823 | -0.142 | -0.0722 | No |
| 118 | Alad | 17830 | -0.142 | -0.0670 | No |
| 119 | Suclg1 | 18170 | -0.154 | -0.0755 | No |
| 120 | Ostc | 18241 | -0.157 | -0.0724 | No |
| 121 | Mdh2 | 18455 | -0.165 | -0.0751 | No |
| 122 | Apex1 | 18718 | -0.177 | -0.0794 | No |
| 123 | Acsl4 | 18854 | -0.183 | -0.0781 | No |
| 124 | S100a10 | 19156 | -0.195 | -0.0833 | No |
| 125 | Hsp90aa1 | 19254 | -0.200 | -0.0797 | No |
| 126 | Aldh9a1 | 19471 | -0.211 | -0.0808 | No |
| 127 | Me1 | 19818 | -0.219 | -0.0870 | No |
| 128 | Psme1 | 20138 | -0.237 | -0.0915 | No |
| 129 | Acat2 | 20450 | -0.256 | -0.0948 | No |
| 130 | Gabarapl1 | 21119 | -0.293 | -0.1120 | No |
| 131 | Eno3 | 21292 | -0.305 | -0.1075 | No |
| 132 | Blvra | 21756 | -0.333 | -0.1144 | No |
| 133 | Fh1 | 22215 | -0.378 | -0.1193 | No |
| 134 | Ehhadh | 22269 | -0.381 | -0.1068 | No |
| 135 | G0s2 | 22532 | -0.420 | -0.1017 | No |
| 136 | Nsdhl | 22899 | -0.476 | -0.0989 | No |
| 137 | Gapdhs | 23430 | -0.634 | -0.0969 | No |
| 138 | Cidea | 23536 | -0.716 | -0.0737 | No |
| 139 | Cpt1a | 23570 | -0.792 | -0.0445 | No |
| 140 | Aoc3 | 23617 | -1.210 | 0.0003 | No |