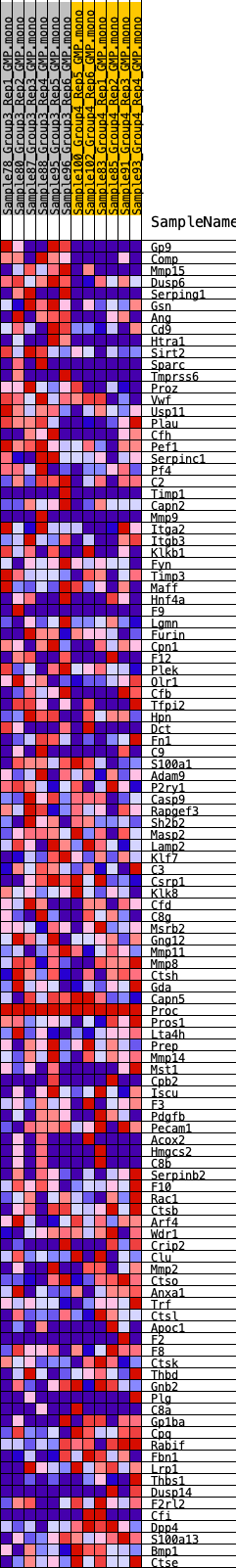
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group3_versus_Group4.GMP.mono_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
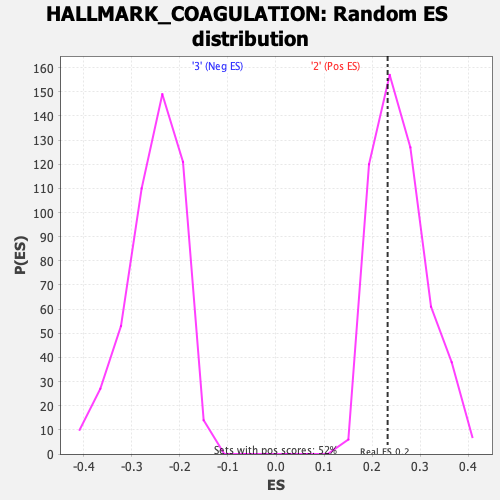
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | 0.2323744 |
| Normalized Enrichment Score (NES) | 0.8989987 |
| Nominal p-value | 0.65697676 |
| FDR q-value | 0.9479186 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gp9 | 167 | 0.805 | 0.0246 | Yes |
| 2 | Comp | 211 | 0.766 | 0.0530 | Yes |
| 3 | Mmp15 | 296 | 0.705 | 0.0773 | Yes |
| 4 | Dusp6 | 488 | 0.634 | 0.0942 | Yes |
| 5 | Serping1 | 715 | 0.566 | 0.1069 | Yes |
| 6 | Gsn | 895 | 0.528 | 0.1201 | Yes |
| 7 | Ang | 930 | 0.520 | 0.1392 | Yes |
| 8 | Cd9 | 966 | 0.512 | 0.1579 | Yes |
| 9 | Htra1 | 984 | 0.508 | 0.1772 | Yes |
| 10 | Sirt2 | 1111 | 0.487 | 0.1910 | Yes |
| 11 | Sparc | 1457 | 0.448 | 0.1940 | Yes |
| 12 | Tmprss6 | 2308 | 0.372 | 0.1725 | Yes |
| 13 | Proz | 2409 | 0.365 | 0.1827 | Yes |
| 14 | Vwf | 2914 | 0.326 | 0.1741 | Yes |
| 15 | Usp11 | 3236 | 0.303 | 0.1724 | Yes |
| 16 | Plau | 3251 | 0.303 | 0.1838 | Yes |
| 17 | Cfh | 3362 | 0.295 | 0.1907 | Yes |
| 18 | Pef1 | 3367 | 0.294 | 0.2022 | Yes |
| 19 | Serpinc1 | 3565 | 0.290 | 0.2052 | Yes |
| 20 | Pf4 | 3687 | 0.281 | 0.2112 | Yes |
| 21 | C2 | 3700 | 0.280 | 0.2217 | Yes |
| 22 | Timp1 | 3929 | 0.271 | 0.2227 | Yes |
| 23 | Capn2 | 4091 | 0.266 | 0.2264 | Yes |
| 24 | Mmp9 | 4292 | 0.254 | 0.2279 | Yes |
| 25 | Itga2 | 4420 | 0.251 | 0.2324 | Yes |
| 26 | Itgb3 | 4683 | 0.239 | 0.2306 | No |
| 27 | Klkb1 | 5073 | 0.220 | 0.2228 | No |
| 28 | Fyn | 5141 | 0.217 | 0.2285 | No |
| 29 | Timp3 | 5850 | 0.187 | 0.2057 | No |
| 30 | Maff | 6001 | 0.181 | 0.2065 | No |
| 31 | Hnf4a | 6016 | 0.180 | 0.2130 | No |
| 32 | F9 | 6266 | 0.171 | 0.2092 | No |
| 33 | Lgmn | 6460 | 0.170 | 0.2077 | No |
| 34 | Furin | 6470 | 0.169 | 0.2140 | No |
| 35 | Cpn1 | 6510 | 0.168 | 0.2189 | No |
| 36 | F12 | 6579 | 0.165 | 0.2226 | No |
| 37 | Plek | 6669 | 0.160 | 0.2251 | No |
| 38 | Olr1 | 6758 | 0.157 | 0.2275 | No |
| 39 | Cfb | 6932 | 0.150 | 0.2261 | No |
| 40 | Tfpi2 | 7043 | 0.147 | 0.2272 | No |
| 41 | Hpn | 7163 | 0.142 | 0.2278 | No |
| 42 | Dct | 7381 | 0.134 | 0.2238 | No |
| 43 | Fn1 | 7502 | 0.130 | 0.2239 | No |
| 44 | C9 | 7630 | 0.126 | 0.2234 | No |
| 45 | S100a1 | 8125 | 0.110 | 0.2068 | No |
| 46 | Adam9 | 8522 | 0.100 | 0.1939 | No |
| 47 | P2ry1 | 8602 | 0.098 | 0.1944 | No |
| 48 | Casp9 | 9341 | 0.077 | 0.1660 | No |
| 49 | Rapgef3 | 9615 | 0.070 | 0.1572 | No |
| 50 | Sh2b2 | 9678 | 0.068 | 0.1572 | No |
| 51 | Masp2 | 9691 | 0.068 | 0.1594 | No |
| 52 | Lamp2 | 9755 | 0.066 | 0.1593 | No |
| 53 | Klf7 | 9846 | 0.063 | 0.1580 | No |
| 54 | C3 | 10111 | 0.057 | 0.1490 | No |
| 55 | Csrp1 | 10458 | 0.047 | 0.1361 | No |
| 56 | Klk8 | 10536 | 0.046 | 0.1347 | No |
| 57 | Cfd | 10736 | 0.040 | 0.1278 | No |
| 58 | C8g | 10844 | 0.038 | 0.1247 | No |
| 59 | Msrb2 | 11122 | 0.030 | 0.1141 | No |
| 60 | Gng12 | 11307 | 0.025 | 0.1073 | No |
| 61 | Mmp11 | 11861 | 0.011 | 0.0842 | No |
| 62 | Mmp8 | 12004 | 0.008 | 0.0785 | No |
| 63 | Ctsh | 12033 | 0.007 | 0.0776 | No |
| 64 | Gda | 12116 | 0.005 | 0.0743 | No |
| 65 | Capn5 | 12294 | 0.000 | 0.0668 | No |
| 66 | Proc | 12340 | 0.000 | 0.0649 | No |
| 67 | Pros1 | 13189 | -0.014 | 0.0294 | No |
| 68 | Lta4h | 13638 | -0.025 | 0.0113 | No |
| 69 | Prep | 13747 | -0.027 | 0.0078 | No |
| 70 | Mmp14 | 14092 | -0.036 | -0.0054 | No |
| 71 | Mst1 | 14191 | -0.038 | -0.0081 | No |
| 72 | Cpb2 | 14560 | -0.048 | -0.0219 | No |
| 73 | Iscu | 14595 | -0.048 | -0.0214 | No |
| 74 | F3 | 14720 | -0.052 | -0.0246 | No |
| 75 | Pdgfb | 15350 | -0.069 | -0.0487 | No |
| 76 | Pecam1 | 15565 | -0.074 | -0.0548 | No |
| 77 | Acox2 | 15586 | -0.075 | -0.0528 | No |
| 78 | Hmgcs2 | 15593 | -0.075 | -0.0501 | No |
| 79 | C8b | 15594 | -0.075 | -0.0471 | No |
| 80 | Serpinb2 | 15877 | -0.082 | -0.0559 | No |
| 81 | F10 | 16233 | -0.093 | -0.0673 | No |
| 82 | Rac1 | 16562 | -0.101 | -0.0773 | No |
| 83 | Ctsb | 17118 | -0.119 | -0.0962 | No |
| 84 | Arf4 | 17789 | -0.140 | -0.1192 | No |
| 85 | Wdr1 | 17871 | -0.143 | -0.1170 | No |
| 86 | Crip2 | 17963 | -0.147 | -0.1150 | No |
| 87 | Clu | 18350 | -0.161 | -0.1251 | No |
| 88 | Mmp2 | 18546 | -0.169 | -0.1267 | No |
| 89 | Ctso | 18840 | -0.182 | -0.1320 | No |
| 90 | Anxa1 | 18936 | -0.186 | -0.1287 | No |
| 91 | Trf | 18939 | -0.186 | -0.1215 | No |
| 92 | Ctsl | 19089 | -0.193 | -0.1202 | No |
| 93 | Apoc1 | 19594 | -0.215 | -0.1332 | No |
| 94 | F2 | 19692 | -0.219 | -0.1287 | No |
| 95 | F8 | 20072 | -0.233 | -0.1356 | No |
| 96 | Ctsk | 20157 | -0.238 | -0.1298 | No |
| 97 | Thbd | 20168 | -0.239 | -0.1208 | No |
| 98 | Gnb2 | 20616 | -0.262 | -0.1295 | No |
| 99 | Plg | 21067 | -0.288 | -0.1373 | No |
| 100 | C8a | 21166 | -0.297 | -0.1297 | No |
| 101 | Gp1ba | 21325 | -0.307 | -0.1243 | No |
| 102 | Cpq | 21566 | -0.318 | -0.1220 | No |
| 103 | Rabif | 21693 | -0.327 | -0.1145 | No |
| 104 | Fbn1 | 21920 | -0.347 | -0.1104 | No |
| 105 | Lrp1 | 22380 | -0.396 | -0.1143 | No |
| 106 | Thbs1 | 22742 | -0.446 | -0.1120 | No |
| 107 | Dusp14 | 23001 | -0.494 | -0.1035 | No |
| 108 | F2rl2 | 23064 | -0.500 | -0.0864 | No |
| 109 | Cfi | 23151 | -0.520 | -0.0696 | No |
| 110 | Dpp4 | 23167 | -0.524 | -0.0495 | No |
| 111 | S100a13 | 23211 | -0.540 | -0.0301 | No |
| 112 | Bmp1 | 23312 | -0.575 | -0.0116 | No |
| 113 | Ctse | 23424 | -0.631 | 0.0085 | No |