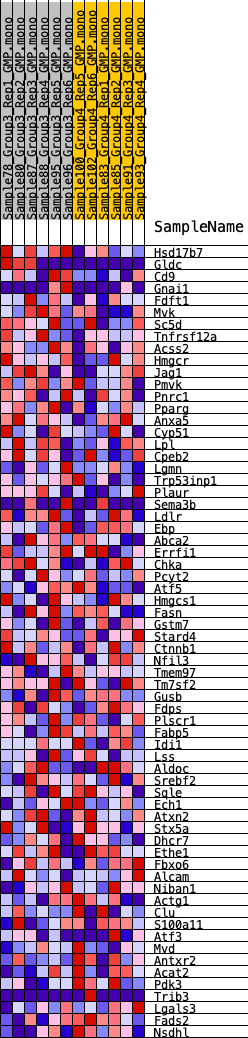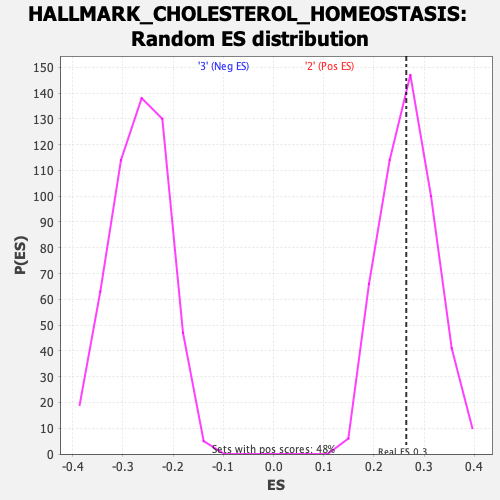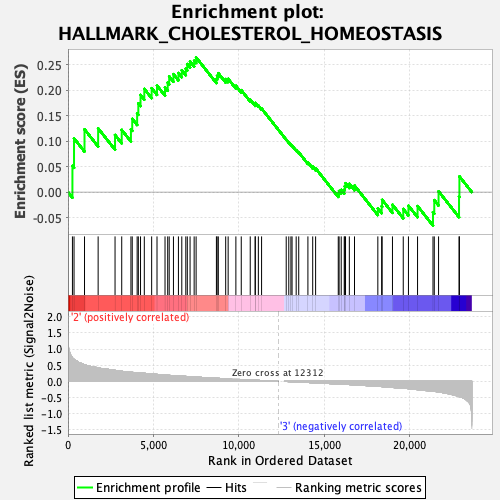
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group3_versus_Group4.GMP.mono_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | 0.26446855 |
| Normalized Enrichment Score (NES) | 0.9881579 |
| Nominal p-value | 0.50619835 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.995 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hsd17b7 | 259 | 0.731 | 0.0515 | Yes |
| 2 | Gldc | 353 | 0.679 | 0.1055 | Yes |
| 3 | Cd9 | 966 | 0.512 | 0.1233 | Yes |
| 4 | Gnai1 | 1763 | 0.418 | 0.1251 | Yes |
| 5 | Fdft1 | 2752 | 0.339 | 0.1122 | Yes |
| 6 | Mvk | 3143 | 0.310 | 0.1221 | Yes |
| 7 | Sc5d | 3682 | 0.282 | 0.1233 | Yes |
| 8 | Tnfrsf12a | 3758 | 0.277 | 0.1438 | Yes |
| 9 | Acss2 | 4045 | 0.269 | 0.1546 | Yes |
| 10 | Hmgcr | 4115 | 0.264 | 0.1743 | Yes |
| 11 | Jag1 | 4243 | 0.256 | 0.1908 | Yes |
| 12 | Pmvk | 4464 | 0.248 | 0.2026 | Yes |
| 13 | Pnrc1 | 4894 | 0.229 | 0.2040 | Yes |
| 14 | Pparg | 5207 | 0.214 | 0.2091 | Yes |
| 15 | Anxa5 | 5676 | 0.193 | 0.2058 | Yes |
| 16 | Cyp51 | 5833 | 0.187 | 0.2151 | Yes |
| 17 | Lpl | 5920 | 0.184 | 0.2272 | Yes |
| 18 | Cpeb2 | 6169 | 0.175 | 0.2316 | Yes |
| 19 | Lgmn | 6460 | 0.170 | 0.2338 | Yes |
| 20 | Trp53inp1 | 6657 | 0.161 | 0.2393 | Yes |
| 21 | Plaur | 6892 | 0.152 | 0.2423 | Yes |
| 22 | Sema3b | 6979 | 0.150 | 0.2514 | Yes |
| 23 | Ldlr | 7141 | 0.143 | 0.2568 | Yes |
| 24 | Ebp | 7380 | 0.134 | 0.2582 | Yes |
| 25 | Abca2 | 7495 | 0.130 | 0.2645 | Yes |
| 26 | Errfi1 | 8682 | 0.095 | 0.2222 | No |
| 27 | Chka | 8723 | 0.094 | 0.2285 | No |
| 28 | Pcyt2 | 8794 | 0.092 | 0.2334 | No |
| 29 | Atf5 | 9225 | 0.080 | 0.2220 | No |
| 30 | Hmgcs1 | 9369 | 0.076 | 0.2224 | No |
| 31 | Fasn | 9821 | 0.064 | 0.2087 | No |
| 32 | Gstm7 | 10136 | 0.056 | 0.2002 | No |
| 33 | Stard4 | 10661 | 0.042 | 0.1815 | No |
| 34 | Ctnnb1 | 10952 | 0.035 | 0.1722 | No |
| 35 | Nfil3 | 10954 | 0.035 | 0.1751 | No |
| 36 | Tmem97 | 11136 | 0.029 | 0.1699 | No |
| 37 | Tm7sf2 | 11325 | 0.025 | 0.1641 | No |
| 38 | Gusb | 12766 | -0.003 | 0.1032 | No |
| 39 | Fdps | 12916 | -0.007 | 0.0974 | No |
| 40 | Plscr1 | 13041 | -0.010 | 0.0930 | No |
| 41 | Fabp5 | 13108 | -0.012 | 0.0912 | No |
| 42 | Idi1 | 13349 | -0.017 | 0.0825 | No |
| 43 | Lss | 13498 | -0.021 | 0.0781 | No |
| 44 | Aldoc | 14035 | -0.035 | 0.0583 | No |
| 45 | Srebf2 | 14316 | -0.042 | 0.0499 | No |
| 46 | Sqle | 14486 | -0.046 | 0.0467 | No |
| 47 | Ech1 | 15811 | -0.080 | -0.0027 | No |
| 48 | Atxn2 | 15860 | -0.082 | 0.0023 | No |
| 49 | Stx5a | 15984 | -0.086 | 0.0044 | No |
| 50 | Dhcr7 | 16160 | -0.091 | 0.0047 | No |
| 51 | Ethe1 | 16191 | -0.092 | 0.0113 | No |
| 52 | Fbxo6 | 16236 | -0.093 | 0.0174 | No |
| 53 | Alcam | 16459 | -0.098 | 0.0164 | No |
| 54 | Niban1 | 16761 | -0.107 | 0.0128 | No |
| 55 | Actg1 | 18126 | -0.152 | -0.0321 | No |
| 56 | Clu | 18350 | -0.161 | -0.0279 | No |
| 57 | S100a11 | 18377 | -0.162 | -0.0152 | No |
| 58 | Atf3 | 18982 | -0.188 | -0.0248 | No |
| 59 | Mvd | 19613 | -0.216 | -0.0331 | No |
| 60 | Antxr2 | 19917 | -0.225 | -0.0268 | No |
| 61 | Acat2 | 20450 | -0.256 | -0.0275 | No |
| 62 | Pdk3 | 21353 | -0.310 | -0.0393 | No |
| 63 | Trib3 | 21428 | -0.313 | -0.0158 | No |
| 64 | Lgals3 | 21679 | -0.326 | 0.0015 | No |
| 65 | Fads2 | 22877 | -0.471 | -0.0090 | No |
| 66 | Nsdhl | 22899 | -0.476 | 0.0308 | No |