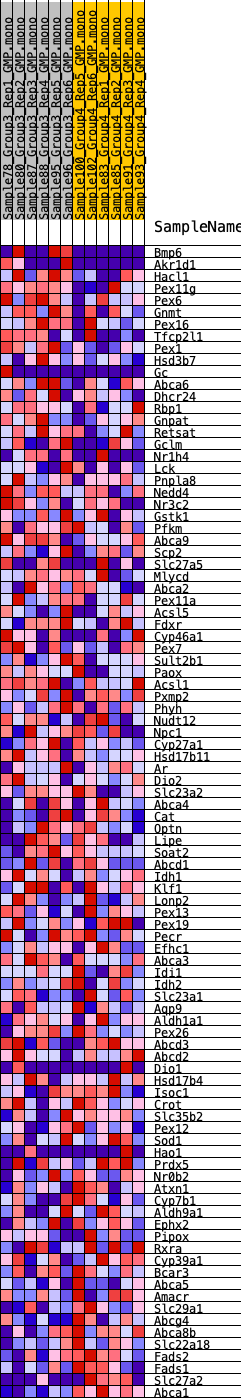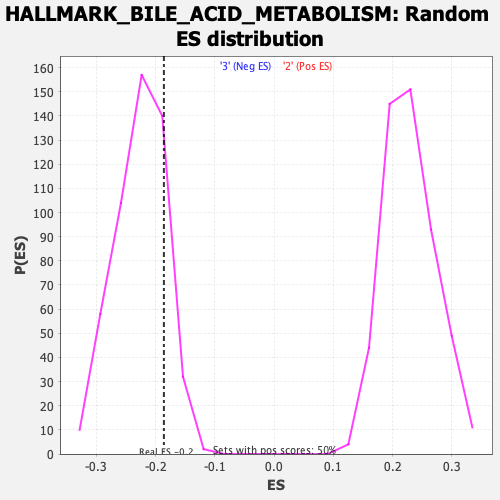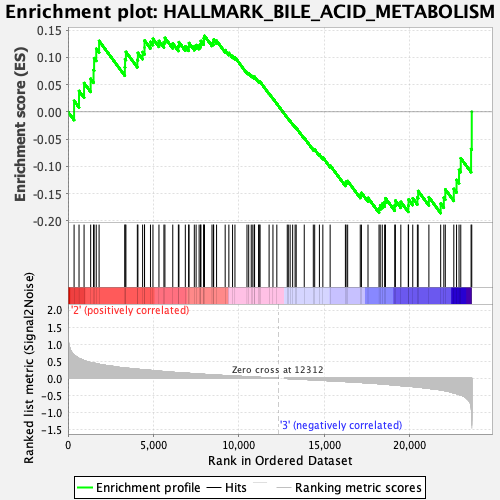
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group3_versus_Group4.GMP.mono_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.18605156 |
| Normalized Enrichment Score (NES) | -0.8224119 |
| Nominal p-value | 0.84691846 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Bmp6 | 356 | 0.679 | 0.0204 | No |
| 2 | Akr1d1 | 645 | 0.579 | 0.0384 | No |
| 3 | Hacl1 | 940 | 0.517 | 0.0530 | No |
| 4 | Pex11g | 1326 | 0.461 | 0.0607 | No |
| 5 | Pex6 | 1495 | 0.446 | 0.0769 | No |
| 6 | Gnmt | 1528 | 0.443 | 0.0987 | No |
| 7 | Pex16 | 1652 | 0.429 | 0.1160 | No |
| 8 | Tfcp2l1 | 1819 | 0.410 | 0.1303 | No |
| 9 | Pex1 | 3326 | 0.297 | 0.0819 | No |
| 10 | Hsd3b7 | 3339 | 0.296 | 0.0969 | No |
| 11 | Gc | 3388 | 0.293 | 0.1102 | No |
| 12 | Abca6 | 4056 | 0.268 | 0.0958 | No |
| 13 | Dhcr24 | 4090 | 0.266 | 0.1083 | No |
| 14 | Rbp1 | 4365 | 0.253 | 0.1099 | No |
| 15 | Gnpat | 4476 | 0.247 | 0.1182 | No |
| 16 | Retsat | 4478 | 0.247 | 0.1311 | No |
| 17 | Gclm | 4825 | 0.233 | 0.1286 | No |
| 18 | Nr1h4 | 4970 | 0.225 | 0.1342 | No |
| 19 | Lck | 5321 | 0.209 | 0.1303 | No |
| 20 | Pnpla8 | 5612 | 0.196 | 0.1282 | No |
| 21 | Nedd4 | 5670 | 0.194 | 0.1359 | No |
| 22 | Nr3c2 | 6129 | 0.176 | 0.1257 | No |
| 23 | Gstk1 | 6457 | 0.170 | 0.1207 | No |
| 24 | Pfkm | 6490 | 0.169 | 0.1281 | No |
| 25 | Abca9 | 6863 | 0.153 | 0.1203 | No |
| 26 | Scp2 | 7058 | 0.147 | 0.1197 | No |
| 27 | Slc27a5 | 7080 | 0.145 | 0.1265 | No |
| 28 | Mlycd | 7390 | 0.134 | 0.1203 | No |
| 29 | Abca2 | 7495 | 0.130 | 0.1227 | No |
| 30 | Pex11a | 7671 | 0.125 | 0.1218 | No |
| 31 | Acsl5 | 7756 | 0.122 | 0.1246 | No |
| 32 | Fdxr | 7774 | 0.122 | 0.1303 | No |
| 33 | Cyp46a1 | 7938 | 0.116 | 0.1294 | No |
| 34 | Pex7 | 7945 | 0.116 | 0.1352 | No |
| 35 | Sult2b1 | 7985 | 0.115 | 0.1396 | No |
| 36 | Paox | 8424 | 0.103 | 0.1264 | No |
| 37 | Acsl1 | 8507 | 0.100 | 0.1281 | No |
| 38 | Pxmp2 | 8521 | 0.100 | 0.1328 | No |
| 39 | Phyh | 8692 | 0.095 | 0.1306 | No |
| 40 | Nudt12 | 9195 | 0.080 | 0.1134 | No |
| 41 | Npc1 | 9411 | 0.075 | 0.1082 | No |
| 42 | Cyp27a1 | 9627 | 0.070 | 0.1027 | No |
| 43 | Hsd17b11 | 9767 | 0.066 | 0.1002 | No |
| 44 | Ar | 10471 | 0.047 | 0.0728 | No |
| 45 | Dio2 | 10571 | 0.045 | 0.0710 | No |
| 46 | Slc23a2 | 10720 | 0.041 | 0.0668 | No |
| 47 | Abca4 | 10813 | 0.038 | 0.0649 | No |
| 48 | Cat | 10900 | 0.036 | 0.0631 | No |
| 49 | Optn | 10916 | 0.036 | 0.0644 | No |
| 50 | Lipe | 11149 | 0.029 | 0.0560 | No |
| 51 | Soat2 | 11191 | 0.028 | 0.0558 | No |
| 52 | Abcd1 | 11238 | 0.027 | 0.0552 | No |
| 53 | Idh1 | 11765 | 0.014 | 0.0336 | No |
| 54 | Klf1 | 11990 | 0.008 | 0.0245 | No |
| 55 | Lonp2 | 12217 | 0.002 | 0.0150 | No |
| 56 | Pex13 | 12827 | -0.004 | -0.0106 | No |
| 57 | Pex19 | 12888 | -0.006 | -0.0129 | No |
| 58 | Pecr | 12995 | -0.009 | -0.0169 | No |
| 59 | Efhc1 | 13143 | -0.012 | -0.0225 | No |
| 60 | Abca3 | 13286 | -0.016 | -0.0277 | No |
| 61 | Idi1 | 13349 | -0.017 | -0.0295 | No |
| 62 | Idh2 | 13821 | -0.030 | -0.0479 | No |
| 63 | Slc23a1 | 14351 | -0.042 | -0.0682 | No |
| 64 | Aqp9 | 14432 | -0.044 | -0.0693 | No |
| 65 | Aldh1a1 | 14708 | -0.052 | -0.0783 | No |
| 66 | Pex26 | 14909 | -0.057 | -0.0838 | No |
| 67 | Abcd3 | 15336 | -0.068 | -0.0983 | No |
| 68 | Abcd2 | 16228 | -0.093 | -0.1313 | No |
| 69 | Dio1 | 16267 | -0.093 | -0.1280 | No |
| 70 | Hsd17b4 | 16361 | -0.095 | -0.1270 | No |
| 71 | Isoc1 | 17098 | -0.118 | -0.1521 | No |
| 72 | Crot | 17167 | -0.120 | -0.1487 | No |
| 73 | Slc35b2 | 17551 | -0.133 | -0.1580 | No |
| 74 | Pex12 | 18196 | -0.155 | -0.1773 | Yes |
| 75 | Sod1 | 18264 | -0.158 | -0.1719 | Yes |
| 76 | Hao1 | 18385 | -0.162 | -0.1685 | Yes |
| 77 | Prdx5 | 18524 | -0.168 | -0.1655 | Yes |
| 78 | Nr0b2 | 18573 | -0.170 | -0.1587 | Yes |
| 79 | Atxn1 | 19116 | -0.194 | -0.1716 | Yes |
| 80 | Cyp7b1 | 19153 | -0.195 | -0.1629 | Yes |
| 81 | Aldh9a1 | 19471 | -0.211 | -0.1653 | Yes |
| 82 | Ephx2 | 19907 | -0.225 | -0.1721 | Yes |
| 83 | Pipox | 19923 | -0.225 | -0.1609 | Yes |
| 84 | Rxra | 20169 | -0.239 | -0.1588 | Yes |
| 85 | Cyp39a1 | 20440 | -0.255 | -0.1570 | Yes |
| 86 | Bcar3 | 20484 | -0.257 | -0.1454 | Yes |
| 87 | Abca5 | 21113 | -0.292 | -0.1568 | Yes |
| 88 | Amacr | 21803 | -0.336 | -0.1685 | Yes |
| 89 | Slc29a1 | 21987 | -0.353 | -0.1577 | Yes |
| 90 | Abcg4 | 22073 | -0.361 | -0.1425 | Yes |
| 91 | Abca8b | 22571 | -0.422 | -0.1415 | Yes |
| 92 | Slc22a18 | 22731 | -0.444 | -0.1250 | Yes |
| 93 | Fads2 | 22877 | -0.471 | -0.1065 | Yes |
| 94 | Fads1 | 22978 | -0.489 | -0.0852 | Yes |
| 95 | Slc27a2 | 23582 | -0.817 | -0.0681 | Yes |
| 96 | Abca1 | 23621 | -1.335 | 0.0001 | Yes |