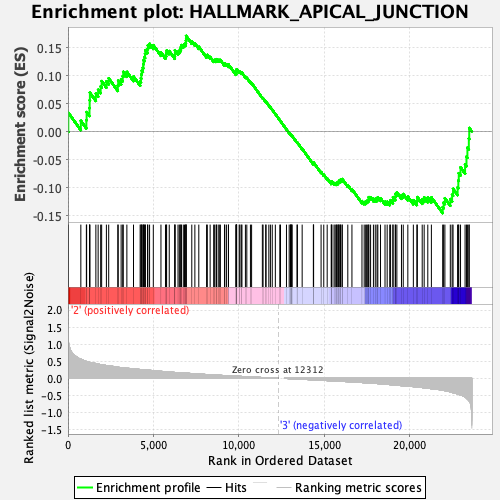
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group3_versus_Group4.GMP.mono_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_APICAL_JUNCTION |
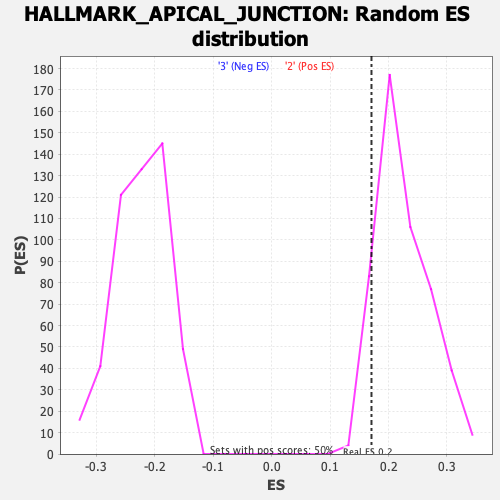
| Enrichment Score (ES) | 0.17087364 |
| Normalized Enrichment Score (NES) | 0.75899374 |
| Nominal p-value | 0.9070707 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tmem8b | 19 | 1.135 | 0.0336 | Yes |
| 2 | Rras | 751 | 0.557 | 0.0193 | Yes |
| 3 | Fscn1 | 1074 | 0.494 | 0.0206 | Yes |
| 4 | Myl9 | 1090 | 0.491 | 0.0348 | Yes |
| 5 | Itgb4 | 1258 | 0.472 | 0.0420 | Yes |
| 6 | Myh10 | 1268 | 0.469 | 0.0558 | Yes |
| 7 | Inppl1 | 1281 | 0.466 | 0.0694 | Yes |
| 8 | Cadm3 | 1630 | 0.433 | 0.0677 | Yes |
| 9 | Gnai1 | 1763 | 0.418 | 0.0748 | Yes |
| 10 | Actn3 | 1911 | 0.400 | 0.0806 | Yes |
| 11 | Adam15 | 1969 | 0.393 | 0.0901 | Yes |
| 12 | Crat | 2246 | 0.374 | 0.0897 | Yes |
| 13 | Thy1 | 2381 | 0.368 | 0.0951 | Yes |
| 14 | Vwf | 2914 | 0.326 | 0.0823 | Yes |
| 15 | Icam2 | 2937 | 0.324 | 0.0912 | Yes |
| 16 | Lama3 | 3108 | 0.313 | 0.0934 | Yes |
| 17 | Adamts5 | 3201 | 0.307 | 0.0988 | Yes |
| 18 | Thbs3 | 3232 | 0.304 | 0.1067 | Yes |
| 19 | Cd276 | 3444 | 0.292 | 0.1066 | Yes |
| 20 | Tspan4 | 3832 | 0.273 | 0.0984 | Yes |
| 21 | Nlgn2 | 4221 | 0.257 | 0.0896 | Yes |
| 22 | Nrtn | 4268 | 0.255 | 0.0954 | Yes |
| 23 | Mmp9 | 4292 | 0.254 | 0.1021 | Yes |
| 24 | Cdh3 | 4315 | 0.254 | 0.1089 | Yes |
| 25 | Cdk8 | 4379 | 0.253 | 0.1138 | Yes |
| 26 | Evl | 4408 | 0.251 | 0.1203 | Yes |
| 27 | Itga2 | 4420 | 0.251 | 0.1274 | Yes |
| 28 | Myh9 | 4471 | 0.248 | 0.1328 | Yes |
| 29 | Map4k2 | 4508 | 0.246 | 0.1387 | Yes |
| 30 | Mpzl1 | 4527 | 0.245 | 0.1453 | Yes |
| 31 | Jam3 | 4652 | 0.241 | 0.1474 | Yes |
| 32 | Adam23 | 4671 | 0.239 | 0.1538 | Yes |
| 33 | Rhof | 4772 | 0.235 | 0.1567 | Yes |
| 34 | Actn2 | 4991 | 0.224 | 0.1542 | Yes |
| 35 | Slc30a3 | 5438 | 0.204 | 0.1413 | Yes |
| 36 | Actb | 5703 | 0.192 | 0.1359 | Yes |
| 37 | Dhx16 | 5749 | 0.190 | 0.1398 | Yes |
| 38 | Ikbkg | 5769 | 0.190 | 0.1447 | Yes |
| 39 | Akt2 | 5923 | 0.184 | 0.1438 | Yes |
| 40 | Plcg1 | 6248 | 0.171 | 0.1351 | Yes |
| 41 | Tro | 6256 | 0.171 | 0.1400 | Yes |
| 42 | Col9a1 | 6257 | 0.171 | 0.1452 | Yes |
| 43 | Nectin2 | 6436 | 0.171 | 0.1428 | Yes |
| 44 | Cldn15 | 6512 | 0.168 | 0.1447 | Yes |
| 45 | Nectin1 | 6566 | 0.165 | 0.1475 | Yes |
| 46 | Nexn | 6605 | 0.164 | 0.1508 | Yes |
| 47 | Zyx | 6636 | 0.162 | 0.1544 | Yes |
| 48 | Tubg1 | 6745 | 0.157 | 0.1546 | Yes |
| 49 | Baiap2 | 6815 | 0.154 | 0.1563 | Yes |
| 50 | Cdh6 | 6888 | 0.152 | 0.1579 | Yes |
| 51 | Src | 6898 | 0.152 | 0.1621 | Yes |
| 52 | Nectin4 | 6908 | 0.152 | 0.1663 | Yes |
| 53 | Cx3cl1 | 6910 | 0.151 | 0.1709 | Yes |
| 54 | Ldlrap1 | 7242 | 0.139 | 0.1610 | No |
| 55 | Egfr | 7422 | 0.133 | 0.1574 | No |
| 56 | Vcam1 | 7652 | 0.125 | 0.1514 | No |
| 57 | Hras | 8107 | 0.111 | 0.1354 | No |
| 58 | Gamt | 8150 | 0.110 | 0.1369 | No |
| 59 | Nfasc | 8315 | 0.107 | 0.1332 | No |
| 60 | Adam9 | 8522 | 0.100 | 0.1274 | No |
| 61 | Wnk4 | 8579 | 0.098 | 0.1280 | No |
| 62 | Mapk11 | 8666 | 0.095 | 0.1272 | No |
| 63 | Icam4 | 8689 | 0.095 | 0.1292 | No |
| 64 | Cercam | 8786 | 0.092 | 0.1279 | No |
| 65 | Tsc1 | 8852 | 0.090 | 0.1278 | No |
| 66 | Actg2 | 8915 | 0.088 | 0.1279 | No |
| 67 | Vcl | 9155 | 0.081 | 0.1201 | No |
| 68 | Kcnh2 | 9172 | 0.081 | 0.1219 | No |
| 69 | Amh | 9268 | 0.079 | 0.1202 | No |
| 70 | Rasa1 | 9383 | 0.075 | 0.1176 | No |
| 71 | Epb41l2 | 9389 | 0.075 | 0.1197 | No |
| 72 | Mapk14 | 9824 | 0.064 | 0.1032 | No |
| 73 | Pals1 | 9842 | 0.063 | 0.1044 | No |
| 74 | Cdh4 | 9844 | 0.063 | 0.1062 | No |
| 75 | Nf2 | 9853 | 0.063 | 0.1078 | No |
| 76 | Pten | 9862 | 0.063 | 0.1094 | No |
| 77 | Nrap | 9865 | 0.063 | 0.1112 | No |
| 78 | Nectin3 | 10013 | 0.059 | 0.1067 | No |
| 79 | Pkd1 | 10055 | 0.058 | 0.1067 | No |
| 80 | Sirpa | 10137 | 0.056 | 0.1050 | No |
| 81 | Amigo1 | 10176 | 0.055 | 0.1050 | No |
| 82 | Ywhah | 10379 | 0.049 | 0.0979 | No |
| 83 | Vasp | 10461 | 0.047 | 0.0959 | No |
| 84 | Stx4a | 10673 | 0.042 | 0.0882 | No |
| 85 | Actn4 | 10741 | 0.040 | 0.0865 | No |
| 86 | Tial1 | 11370 | 0.024 | 0.0604 | No |
| 87 | B4galt1 | 11412 | 0.023 | 0.0594 | No |
| 88 | Dlg1 | 11567 | 0.019 | 0.0534 | No |
| 89 | Irs1 | 11586 | 0.018 | 0.0532 | No |
| 90 | Shc1 | 11749 | 0.014 | 0.0467 | No |
| 91 | Hadh | 11841 | 0.012 | 0.0432 | No |
| 92 | Nf1 | 11949 | 0.009 | 0.0389 | No |
| 93 | Actn1 | 12130 | 0.005 | 0.0314 | No |
| 94 | Dsc1 | 12391 | 0.000 | 0.0203 | No |
| 95 | Flnc | 12432 | 0.000 | 0.0186 | No |
| 96 | Msn | 12781 | -0.003 | 0.0038 | No |
| 97 | Rsu1 | 12940 | -0.007 | -0.0027 | No |
| 98 | Map3k20 | 13012 | -0.009 | -0.0054 | No |
| 99 | Negr1 | 13016 | -0.010 | -0.0053 | No |
| 100 | Sorbs3 | 13056 | -0.010 | -0.0066 | No |
| 101 | Mapk13 | 13058 | -0.011 | -0.0063 | No |
| 102 | Insig1 | 13095 | -0.011 | -0.0075 | No |
| 103 | Cd34 | 13107 | -0.012 | -0.0077 | No |
| 104 | Pfn1 | 13398 | -0.019 | -0.0195 | No |
| 105 | Ptprc | 13421 | -0.019 | -0.0198 | No |
| 106 | Cdh1 | 13699 | -0.026 | -0.0308 | No |
| 107 | Exoc4 | 14357 | -0.043 | -0.0576 | No |
| 108 | Wasl | 14359 | -0.043 | -0.0563 | No |
| 109 | Myl12b | 14369 | -0.043 | -0.0554 | No |
| 110 | Lamb3 | 14806 | -0.055 | -0.0723 | No |
| 111 | Lima1 | 14963 | -0.059 | -0.0772 | No |
| 112 | Vcan | 15163 | -0.065 | -0.0837 | No |
| 113 | Nrxn2 | 15399 | -0.070 | -0.0916 | No |
| 114 | Atp1a3 | 15420 | -0.070 | -0.0904 | No |
| 115 | Vav2 | 15438 | -0.071 | -0.0889 | No |
| 116 | Pecam1 | 15565 | -0.074 | -0.0921 | No |
| 117 | Adra1b | 15631 | -0.076 | -0.0925 | No |
| 118 | Cldn14 | 15698 | -0.077 | -0.0930 | No |
| 119 | Akt3 | 15718 | -0.078 | -0.0915 | No |
| 120 | Gtf2f1 | 15788 | -0.080 | -0.0920 | No |
| 121 | Cnn2 | 15807 | -0.080 | -0.0903 | No |
| 122 | Syk | 15848 | -0.081 | -0.0896 | No |
| 123 | Mdk | 15859 | -0.082 | -0.0875 | No |
| 124 | Cd86 | 15931 | -0.084 | -0.0880 | No |
| 125 | Taok2 | 15942 | -0.084 | -0.0859 | No |
| 126 | Skap2 | 16035 | -0.087 | -0.0872 | No |
| 127 | Arpc2 | 16043 | -0.088 | -0.0848 | No |
| 128 | Gnai2 | 16369 | -0.095 | -0.0958 | No |
| 129 | Speg | 16611 | -0.102 | -0.1029 | No |
| 130 | Pik3cb | 17198 | -0.122 | -0.1242 | No |
| 131 | Itgb1 | 17328 | -0.126 | -0.1259 | No |
| 132 | Arhgef6 | 17415 | -0.129 | -0.1257 | No |
| 133 | Cap1 | 17472 | -0.131 | -0.1241 | No |
| 134 | Crb3 | 17534 | -0.133 | -0.1227 | No |
| 135 | Acta1 | 17591 | -0.134 | -0.1210 | No |
| 136 | Traf1 | 17597 | -0.134 | -0.1171 | No |
| 137 | Tjp1 | 17702 | -0.137 | -0.1174 | No |
| 138 | Ctnna1 | 17868 | -0.143 | -0.1201 | No |
| 139 | Itga3 | 17975 | -0.147 | -0.1202 | No |
| 140 | Pik3r3 | 18078 | -0.151 | -0.1199 | No |
| 141 | Actg1 | 18126 | -0.152 | -0.1173 | No |
| 142 | Cntn1 | 18283 | -0.158 | -0.1192 | No |
| 143 | Mmp2 | 18546 | -0.169 | -0.1252 | No |
| 144 | Sympk | 18672 | -0.175 | -0.1253 | No |
| 145 | Cd274 | 18823 | -0.182 | -0.1261 | No |
| 146 | Itga9 | 18871 | -0.184 | -0.1226 | No |
| 147 | Pard6g | 19009 | -0.189 | -0.1227 | No |
| 148 | Ptk2 | 19015 | -0.189 | -0.1172 | No |
| 149 | Ctnnd1 | 19145 | -0.195 | -0.1167 | No |
| 150 | Cdh8 | 19155 | -0.195 | -0.1112 | No |
| 151 | Cadm2 | 19239 | -0.199 | -0.1087 | No |
| 152 | Calb2 | 19509 | -0.213 | -0.1137 | No |
| 153 | Mvd | 19613 | -0.216 | -0.1116 | No |
| 154 | Icam5 | 19881 | -0.223 | -0.1162 | No |
| 155 | Parva | 20204 | -0.241 | -0.1226 | No |
| 156 | Cdh11 | 20410 | -0.254 | -0.1237 | No |
| 157 | Pbx2 | 20437 | -0.255 | -0.1170 | No |
| 158 | Layn | 20722 | -0.268 | -0.1210 | No |
| 159 | Alox8 | 20837 | -0.277 | -0.1175 | No |
| 160 | Jup | 21049 | -0.287 | -0.1178 | No |
| 161 | Rac2 | 21259 | -0.302 | -0.1176 | No |
| 162 | Fbn1 | 21920 | -0.347 | -0.1352 | No |
| 163 | Lamc2 | 21974 | -0.352 | -0.1268 | No |
| 164 | Col17a1 | 22049 | -0.358 | -0.1190 | No |
| 165 | Sdc3 | 22366 | -0.394 | -0.1206 | No |
| 166 | Nlgn3 | 22471 | -0.408 | -0.1126 | No |
| 167 | Tgfbi | 22526 | -0.420 | -0.1022 | No |
| 168 | Sgce | 22793 | -0.456 | -0.0997 | No |
| 169 | Dsc3 | 22835 | -0.463 | -0.0874 | No |
| 170 | Icam1 | 22862 | -0.467 | -0.0743 | No |
| 171 | Itga10 | 22964 | -0.487 | -0.0639 | No |
| 172 | Shroom2 | 23230 | -0.546 | -0.0586 | No |
| 173 | Bmp1 | 23312 | -0.575 | -0.0446 | No |
| 174 | Col16a1 | 23367 | -0.604 | -0.0286 | No |
| 175 | Amigo2 | 23450 | -0.643 | -0.0126 | No |
| 176 | Slit2 | 23474 | -0.659 | 0.0064 | No |