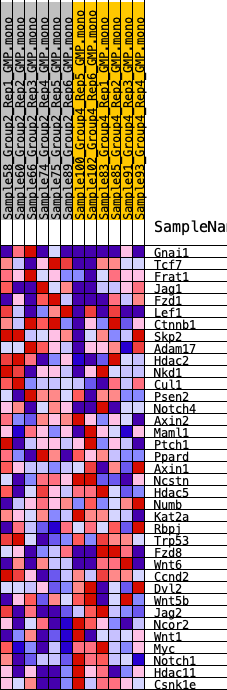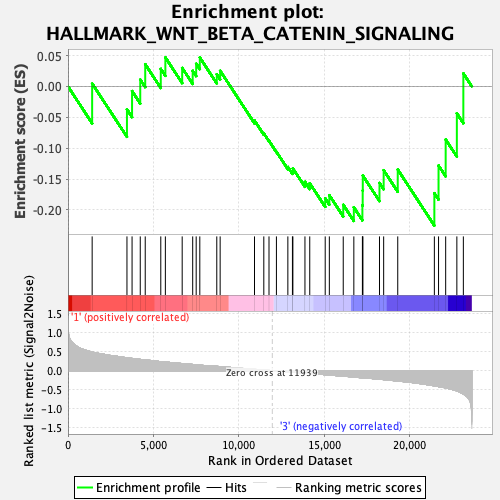
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group2_versus_Group4.GMP.mono_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_WNT_BETA_CATENIN_SIGNALING |
| Enrichment Score (ES) | -0.22536117 |
| Normalized Enrichment Score (NES) | -0.81522083 |
| Nominal p-value | 0.7016129 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gnai1 | 1413 | 0.494 | 0.0046 | No |
| 2 | Tcf7 | 3449 | 0.340 | -0.0372 | No |
| 3 | Frat1 | 3748 | 0.323 | -0.0075 | No |
| 4 | Jag1 | 4227 | 0.297 | 0.0111 | No |
| 5 | Fzd1 | 4518 | 0.284 | 0.0359 | No |
| 6 | Lef1 | 5428 | 0.237 | 0.0284 | No |
| 7 | Ctnnb1 | 5696 | 0.228 | 0.0469 | No |
| 8 | Skp2 | 6680 | 0.189 | 0.0299 | No |
| 9 | Adam17 | 7295 | 0.163 | 0.0252 | No |
| 10 | Hdac2 | 7499 | 0.154 | 0.0368 | No |
| 11 | Nkd1 | 7711 | 0.145 | 0.0468 | No |
| 12 | Cul1 | 8704 | 0.114 | 0.0197 | No |
| 13 | Psen2 | 8901 | 0.107 | 0.0254 | No |
| 14 | Notch4 | 10911 | 0.035 | -0.0552 | No |
| 15 | Axin2 | 11455 | 0.017 | -0.0761 | No |
| 16 | Maml1 | 11759 | 0.006 | -0.0882 | No |
| 17 | Ptch1 | 12193 | -0.005 | -0.1058 | No |
| 18 | Ppard | 12862 | -0.029 | -0.1304 | No |
| 19 | Axin1 | 13130 | -0.038 | -0.1368 | No |
| 20 | Ncstn | 13156 | -0.039 | -0.1328 | No |
| 21 | Hdac5 | 13862 | -0.063 | -0.1545 | No |
| 22 | Numb | 14143 | -0.073 | -0.1568 | No |
| 23 | Kat2a | 15047 | -0.106 | -0.1812 | No |
| 24 | Rbpj | 15289 | -0.116 | -0.1762 | No |
| 25 | Trp53 | 16102 | -0.145 | -0.1916 | No |
| 26 | Fzd8 | 16722 | -0.170 | -0.1957 | Yes |
| 27 | Wnt6 | 17223 | -0.187 | -0.1925 | Yes |
| 28 | Ccnd2 | 17240 | -0.187 | -0.1687 | Yes |
| 29 | Dvl2 | 17246 | -0.187 | -0.1444 | Yes |
| 30 | Wnt5b | 18223 | -0.225 | -0.1563 | Yes |
| 31 | Jag2 | 18467 | -0.236 | -0.1357 | Yes |
| 32 | Ncor2 | 19291 | -0.275 | -0.1346 | Yes |
| 33 | Wnt1 | 21433 | -0.401 | -0.1729 | Yes |
| 34 | Myc | 21678 | -0.422 | -0.1281 | Yes |
| 35 | Notch1 | 22095 | -0.457 | -0.0860 | Yes |
| 36 | Hdac11 | 22749 | -0.536 | -0.0436 | Yes |
| 37 | Csnk1e | 23129 | -0.617 | 0.0210 | Yes |