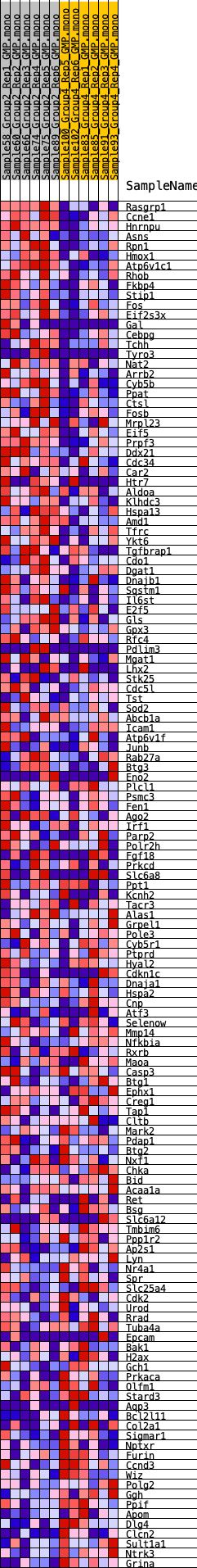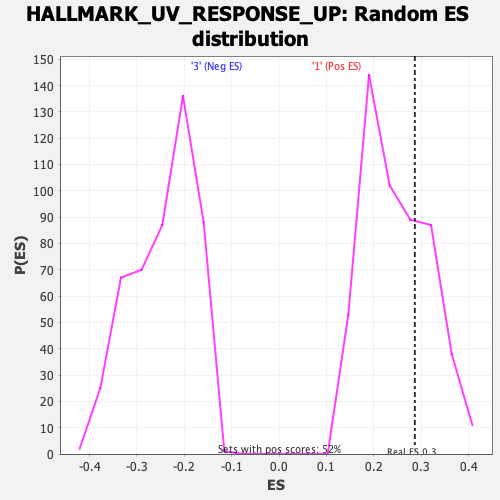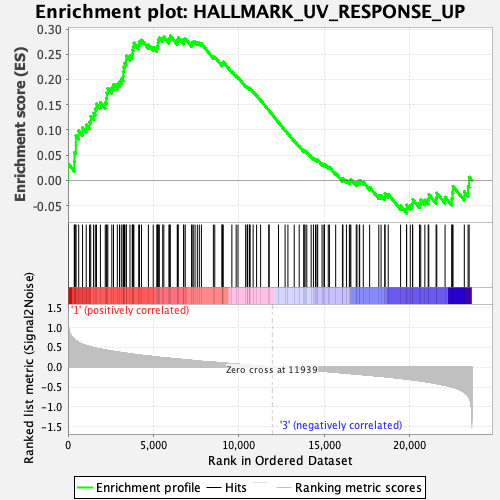
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group2_versus_Group4.GMP.mono_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_UV_RESPONSE_UP |
| Enrichment Score (ES) | 0.28644022 |
| Normalized Enrichment Score (NES) | 1.153078 |
| Nominal p-value | 0.29580152 |
| FDR q-value | 0.47527146 |
| FWER p-Value | 0.935 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Rasgrp1 | 6 | 1.215 | 0.0332 | Yes |
| 2 | Ccne1 | 365 | 0.693 | 0.0371 | Yes |
| 3 | Hnrnpu | 380 | 0.687 | 0.0555 | Yes |
| 4 | Asns | 459 | 0.660 | 0.0703 | Yes |
| 5 | Rpn1 | 460 | 0.660 | 0.0886 | Yes |
| 6 | Hmox1 | 626 | 0.610 | 0.0983 | Yes |
| 7 | Atp6v1c1 | 852 | 0.567 | 0.1044 | Yes |
| 8 | Rhob | 1065 | 0.536 | 0.1101 | Yes |
| 9 | Fkbp4 | 1262 | 0.512 | 0.1159 | Yes |
| 10 | Stip1 | 1328 | 0.501 | 0.1270 | Yes |
| 11 | Fos | 1497 | 0.484 | 0.1332 | Yes |
| 12 | Eif2s3x | 1596 | 0.471 | 0.1420 | Yes |
| 13 | Gal | 1669 | 0.464 | 0.1517 | Yes |
| 14 | Cebpg | 1895 | 0.448 | 0.1545 | Yes |
| 15 | Tchh | 2179 | 0.426 | 0.1542 | Yes |
| 16 | Tyro3 | 2242 | 0.420 | 0.1631 | Yes |
| 17 | Nat2 | 2273 | 0.418 | 0.1734 | Yes |
| 18 | Arrb2 | 2329 | 0.412 | 0.1824 | Yes |
| 19 | Cyb5b | 2564 | 0.396 | 0.1834 | Yes |
| 20 | Ppat | 2662 | 0.391 | 0.1900 | Yes |
| 21 | Ctsl | 2885 | 0.378 | 0.1910 | Yes |
| 22 | Fosb | 3013 | 0.367 | 0.1957 | Yes |
| 23 | Mrpl23 | 3116 | 0.361 | 0.2013 | Yes |
| 24 | Eif5 | 3224 | 0.355 | 0.2066 | Yes |
| 25 | Prpf3 | 3238 | 0.354 | 0.2158 | Yes |
| 26 | Ddx21 | 3263 | 0.352 | 0.2244 | Yes |
| 27 | Cdc34 | 3305 | 0.348 | 0.2323 | Yes |
| 28 | Car2 | 3404 | 0.343 | 0.2376 | Yes |
| 29 | Htr7 | 3407 | 0.343 | 0.2470 | Yes |
| 30 | Aldoa | 3625 | 0.330 | 0.2468 | Yes |
| 31 | Klhdc3 | 3759 | 0.322 | 0.2500 | Yes |
| 32 | Hspa13 | 3774 | 0.321 | 0.2583 | Yes |
| 33 | Amd1 | 3819 | 0.318 | 0.2652 | Yes |
| 34 | Tfrc | 3855 | 0.315 | 0.2724 | Yes |
| 35 | Ykt6 | 4138 | 0.303 | 0.2687 | Yes |
| 36 | Tgfbrap1 | 4180 | 0.300 | 0.2753 | Yes |
| 37 | Cdo1 | 4299 | 0.293 | 0.2783 | Yes |
| 38 | Dgat1 | 4703 | 0.275 | 0.2688 | Yes |
| 39 | Dnajb1 | 4986 | 0.260 | 0.2639 | Yes |
| 40 | Sqstm1 | 5196 | 0.249 | 0.2619 | Yes |
| 41 | Il6st | 5239 | 0.247 | 0.2669 | Yes |
| 42 | E2f5 | 5269 | 0.246 | 0.2725 | Yes |
| 43 | Gls | 5277 | 0.245 | 0.2789 | Yes |
| 44 | Gpx3 | 5343 | 0.242 | 0.2828 | Yes |
| 45 | Rfc4 | 5540 | 0.232 | 0.2809 | Yes |
| 46 | Pdlim3 | 5608 | 0.229 | 0.2843 | Yes |
| 47 | Mgat1 | 5904 | 0.220 | 0.2778 | Yes |
| 48 | Lhx2 | 5948 | 0.217 | 0.2820 | Yes |
| 49 | Stk25 | 5984 | 0.216 | 0.2864 | Yes |
| 50 | Cdc5l | 6392 | 0.200 | 0.2746 | No |
| 51 | Tst | 6411 | 0.199 | 0.2793 | No |
| 52 | Sod2 | 6451 | 0.197 | 0.2831 | No |
| 53 | Abcb1a | 6764 | 0.185 | 0.2749 | No |
| 54 | Icam1 | 6769 | 0.185 | 0.2798 | No |
| 55 | Atp6v1f | 6859 | 0.181 | 0.2810 | No |
| 56 | Junb | 7230 | 0.166 | 0.2699 | No |
| 57 | Rab27a | 7255 | 0.165 | 0.2734 | No |
| 58 | Btg3 | 7326 | 0.162 | 0.2749 | No |
| 59 | Eno2 | 7432 | 0.158 | 0.2748 | No |
| 60 | Plcl1 | 7566 | 0.151 | 0.2733 | No |
| 61 | Psmc3 | 7683 | 0.146 | 0.2724 | No |
| 62 | Fen1 | 7804 | 0.142 | 0.2712 | No |
| 63 | Ago2 | 8505 | 0.122 | 0.2447 | No |
| 64 | Irf1 | 8586 | 0.118 | 0.2446 | No |
| 65 | Parp2 | 9010 | 0.103 | 0.2294 | No |
| 66 | Polr2h | 9017 | 0.103 | 0.2320 | No |
| 67 | Fgf18 | 9047 | 0.101 | 0.2335 | No |
| 68 | Prkcd | 9080 | 0.100 | 0.2349 | No |
| 69 | Slc6a8 | 9587 | 0.082 | 0.2157 | No |
| 70 | Ppt1 | 9838 | 0.072 | 0.2070 | No |
| 71 | Kcnh2 | 9946 | 0.068 | 0.2043 | No |
| 72 | Tacr3 | 10392 | 0.053 | 0.1868 | No |
| 73 | Alas1 | 10480 | 0.050 | 0.1845 | No |
| 74 | Grpel1 | 10500 | 0.049 | 0.1850 | No |
| 75 | Pole3 | 10605 | 0.045 | 0.1819 | No |
| 76 | Cyb5r1 | 10632 | 0.045 | 0.1820 | No |
| 77 | Ptprd | 10649 | 0.044 | 0.1825 | No |
| 78 | Hyal2 | 10832 | 0.037 | 0.1758 | No |
| 79 | Cdkn1c | 11033 | 0.030 | 0.1681 | No |
| 80 | Dnaja1 | 11269 | 0.023 | 0.1587 | No |
| 81 | Hspa2 | 11744 | 0.007 | 0.1387 | No |
| 82 | Cnp | 11768 | 0.006 | 0.1379 | No |
| 83 | Atf3 | 12314 | -0.010 | 0.1150 | No |
| 84 | Selenow | 12701 | -0.023 | 0.0992 | No |
| 85 | Mmp14 | 12866 | -0.029 | 0.0930 | No |
| 86 | Nfkbia | 13235 | -0.042 | 0.0785 | No |
| 87 | Rxrb | 13525 | -0.052 | 0.0676 | No |
| 88 | Maoa | 13795 | -0.061 | 0.0579 | No |
| 89 | Casp3 | 13810 | -0.061 | 0.0589 | No |
| 90 | Btg1 | 13869 | -0.063 | 0.0582 | No |
| 91 | Ephx1 | 13970 | -0.067 | 0.0558 | No |
| 92 | Creg1 | 14225 | -0.076 | 0.0471 | No |
| 93 | Tap1 | 14358 | -0.081 | 0.0437 | No |
| 94 | Cltb | 14479 | -0.086 | 0.0410 | No |
| 95 | Mark2 | 14565 | -0.089 | 0.0398 | No |
| 96 | Pdap1 | 14608 | -0.091 | 0.0405 | No |
| 97 | Btg2 | 14863 | -0.099 | 0.0325 | No |
| 98 | Nxf1 | 14972 | -0.104 | 0.0307 | No |
| 99 | Chka | 15006 | -0.105 | 0.0322 | No |
| 100 | Bid | 15225 | -0.114 | 0.0261 | No |
| 101 | Acaa1a | 15294 | -0.117 | 0.0264 | No |
| 102 | Ret | 15660 | -0.129 | 0.0144 | No |
| 103 | Bsg | 16055 | -0.144 | 0.0016 | No |
| 104 | Slc6a12 | 16084 | -0.145 | 0.0044 | No |
| 105 | Tmbim6 | 16289 | -0.152 | -0.0001 | No |
| 106 | Ppp1r2 | 16459 | -0.158 | -0.0029 | No |
| 107 | Ap2s1 | 16503 | -0.160 | -0.0003 | No |
| 108 | Lyn | 16548 | -0.162 | 0.0023 | No |
| 109 | Nr4a1 | 16863 | -0.175 | -0.0063 | No |
| 110 | Spr | 16905 | -0.177 | -0.0031 | No |
| 111 | Slc25a4 | 17041 | -0.183 | -0.0038 | No |
| 112 | Cdk2 | 17064 | -0.183 | 0.0003 | No |
| 113 | Urod | 17275 | -0.189 | -0.0035 | No |
| 114 | Rrad | 17647 | -0.203 | -0.0137 | No |
| 115 | Tuba4a | 18182 | -0.223 | -0.0302 | No |
| 116 | Epcam | 18314 | -0.229 | -0.0295 | No |
| 117 | Bak1 | 18534 | -0.238 | -0.0323 | No |
| 118 | H2ax | 18546 | -0.238 | -0.0262 | No |
| 119 | Gch1 | 18734 | -0.248 | -0.0273 | No |
| 120 | Prkaca | 19458 | -0.281 | -0.0503 | No |
| 121 | Olfm1 | 19804 | -0.301 | -0.0567 | No |
| 122 | Stard3 | 19812 | -0.302 | -0.0487 | No |
| 123 | Aqp3 | 20024 | -0.312 | -0.0490 | No |
| 124 | Bcl2l11 | 20156 | -0.316 | -0.0459 | No |
| 125 | Col2a1 | 20167 | -0.317 | -0.0376 | No |
| 126 | Sigmar1 | 20566 | -0.340 | -0.0452 | No |
| 127 | Nptxr | 20637 | -0.346 | -0.0386 | No |
| 128 | Furin | 20877 | -0.361 | -0.0388 | No |
| 129 | Ccnd3 | 21072 | -0.374 | -0.0368 | No |
| 130 | Wiz | 21107 | -0.376 | -0.0278 | No |
| 131 | Polg2 | 21534 | -0.409 | -0.0347 | No |
| 132 | Ggh | 21574 | -0.412 | -0.0250 | No |
| 133 | Ppif | 22060 | -0.454 | -0.0331 | No |
| 134 | Apom | 22446 | -0.494 | -0.0359 | No |
| 135 | Dlg4 | 22488 | -0.499 | -0.0239 | No |
| 136 | Clcn2 | 22525 | -0.504 | -0.0115 | No |
| 137 | Sult1a1 | 23185 | -0.633 | -0.0221 | No |
| 138 | Ntrk3 | 23407 | -0.719 | -0.0117 | No |
| 139 | Grina | 23468 | -0.759 | 0.0066 | No |