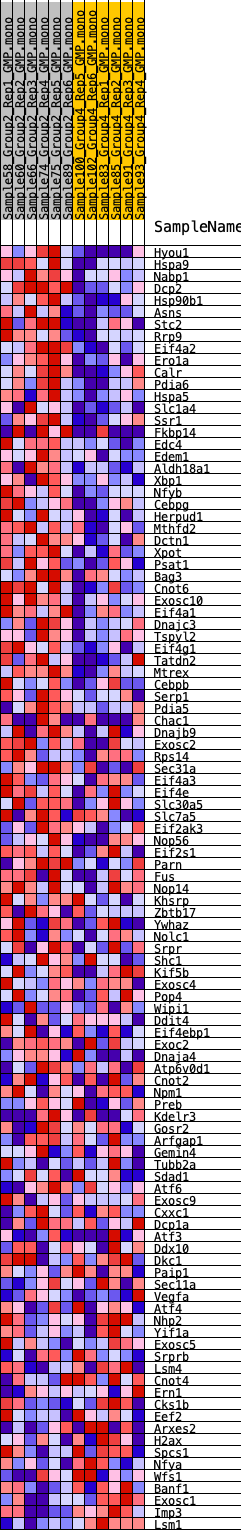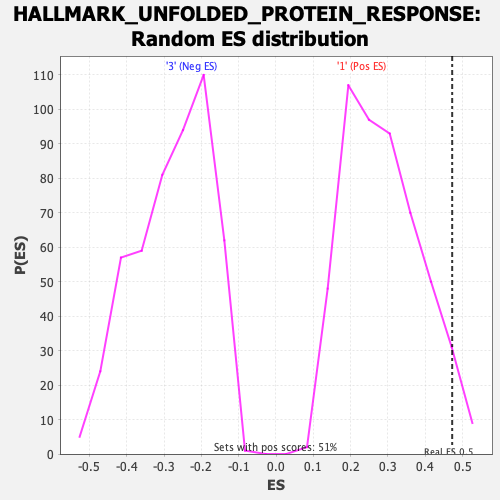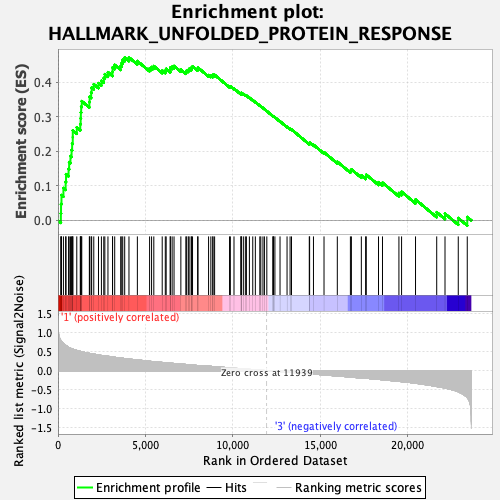
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group2_versus_Group4.GMP.mono_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.47211882 |
| Normalized Enrichment Score (NES) | 1.6506592 |
| Nominal p-value | 0.043392505 |
| FDR q-value | 0.095945746 |
| FWER p-Value | 0.207 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hyou1 | 164 | 0.793 | 0.0207 | Yes |
| 2 | Hspa9 | 174 | 0.786 | 0.0476 | Yes |
| 3 | Nabp1 | 199 | 0.771 | 0.0735 | Yes |
| 4 | Dcp2 | 310 | 0.719 | 0.0938 | Yes |
| 5 | Hsp90b1 | 436 | 0.671 | 0.1118 | Yes |
| 6 | Asns | 459 | 0.660 | 0.1339 | Yes |
| 7 | Stc2 | 604 | 0.614 | 0.1492 | Yes |
| 8 | Rrp9 | 637 | 0.608 | 0.1690 | Yes |
| 9 | Eif4a2 | 712 | 0.592 | 0.1865 | Yes |
| 10 | Ero1a | 776 | 0.580 | 0.2040 | Yes |
| 11 | Calr | 808 | 0.574 | 0.2227 | Yes |
| 12 | Pdia6 | 843 | 0.567 | 0.2410 | Yes |
| 13 | Hspa5 | 845 | 0.567 | 0.2607 | Yes |
| 14 | Slc1a4 | 1073 | 0.535 | 0.2696 | Yes |
| 15 | Ssr1 | 1273 | 0.510 | 0.2789 | Yes |
| 16 | Fkbp14 | 1294 | 0.505 | 0.2957 | Yes |
| 17 | Edc4 | 1307 | 0.503 | 0.3127 | Yes |
| 18 | Edem1 | 1325 | 0.502 | 0.3295 | Yes |
| 19 | Aldh18a1 | 1358 | 0.498 | 0.3454 | Yes |
| 20 | Xbp1 | 1792 | 0.456 | 0.3429 | Yes |
| 21 | Nfyb | 1807 | 0.455 | 0.3582 | Yes |
| 22 | Cebpg | 1895 | 0.448 | 0.3701 | Yes |
| 23 | Herpud1 | 1924 | 0.446 | 0.3844 | Yes |
| 24 | Mthfd2 | 2045 | 0.437 | 0.3945 | Yes |
| 25 | Dctn1 | 2314 | 0.413 | 0.3976 | Yes |
| 26 | Xpot | 2481 | 0.401 | 0.4045 | Yes |
| 27 | Psat1 | 2616 | 0.394 | 0.4125 | Yes |
| 28 | Bag3 | 2677 | 0.389 | 0.4235 | Yes |
| 29 | Cnot6 | 2855 | 0.380 | 0.4292 | Yes |
| 30 | Exosc10 | 3115 | 0.361 | 0.4307 | Yes |
| 31 | Eif4a1 | 3120 | 0.361 | 0.4431 | Yes |
| 32 | Dnajc3 | 3241 | 0.353 | 0.4503 | Yes |
| 33 | Tspyl2 | 3583 | 0.333 | 0.4474 | Yes |
| 34 | Eif4g1 | 3645 | 0.329 | 0.4563 | Yes |
| 35 | Tatdn2 | 3701 | 0.327 | 0.4653 | Yes |
| 36 | Mtrex | 3812 | 0.319 | 0.4718 | Yes |
| 37 | Cebpb | 4056 | 0.307 | 0.4721 | Yes |
| 38 | Serp1 | 4536 | 0.282 | 0.4616 | No |
| 39 | Pdia5 | 5235 | 0.247 | 0.4405 | No |
| 40 | Chac1 | 5348 | 0.242 | 0.4442 | No |
| 41 | Dnajb9 | 5476 | 0.235 | 0.4470 | No |
| 42 | Exosc2 | 5956 | 0.217 | 0.4341 | No |
| 43 | Rps14 | 6132 | 0.208 | 0.4339 | No |
| 44 | Sec31a | 6182 | 0.206 | 0.4390 | No |
| 45 | Eif4a3 | 6416 | 0.198 | 0.4360 | No |
| 46 | Eif4e | 6419 | 0.198 | 0.4428 | No |
| 47 | Slc30a5 | 6523 | 0.193 | 0.4452 | No |
| 48 | Slc7a5 | 6632 | 0.189 | 0.4472 | No |
| 49 | Eif2ak3 | 7022 | 0.174 | 0.4367 | No |
| 50 | Nop56 | 7307 | 0.163 | 0.4303 | No |
| 51 | Eif2s1 | 7353 | 0.161 | 0.4340 | No |
| 52 | Parn | 7448 | 0.157 | 0.4355 | No |
| 53 | Fus | 7494 | 0.155 | 0.4390 | No |
| 54 | Nop14 | 7608 | 0.150 | 0.4394 | No |
| 55 | Khsrp | 7635 | 0.149 | 0.4434 | No |
| 56 | Zbtb17 | 7689 | 0.146 | 0.4463 | No |
| 57 | Ywhaz | 7980 | 0.135 | 0.4386 | No |
| 58 | Nolc1 | 8002 | 0.134 | 0.4424 | No |
| 59 | Srpr | 8606 | 0.118 | 0.4208 | No |
| 60 | Shc1 | 8736 | 0.113 | 0.4193 | No |
| 61 | Kif5b | 8845 | 0.109 | 0.4185 | No |
| 62 | Exosc4 | 8858 | 0.108 | 0.4218 | No |
| 63 | Pop4 | 8951 | 0.105 | 0.4215 | No |
| 64 | Wipi1 | 9804 | 0.074 | 0.3879 | No |
| 65 | Ddit4 | 9851 | 0.071 | 0.3884 | No |
| 66 | Eif4ebp1 | 10059 | 0.064 | 0.3818 | No |
| 67 | Exoc2 | 10460 | 0.050 | 0.3666 | No |
| 68 | Dnaja4 | 10465 | 0.050 | 0.3681 | No |
| 69 | Atp6v0d1 | 10475 | 0.050 | 0.3695 | No |
| 70 | Cnot2 | 10593 | 0.046 | 0.3661 | No |
| 71 | Npm1 | 10707 | 0.042 | 0.3628 | No |
| 72 | Preb | 10768 | 0.040 | 0.3616 | No |
| 73 | Kdelr3 | 10939 | 0.034 | 0.3556 | No |
| 74 | Gosr2 | 11131 | 0.028 | 0.3484 | No |
| 75 | Arfgap1 | 11279 | 0.023 | 0.3430 | No |
| 76 | Gemin4 | 11535 | 0.013 | 0.3326 | No |
| 77 | Tubb2a | 11591 | 0.012 | 0.3307 | No |
| 78 | Sdad1 | 11714 | 0.007 | 0.3257 | No |
| 79 | Atf6 | 11798 | 0.005 | 0.3224 | No |
| 80 | Exosc9 | 11935 | 0.000 | 0.3166 | No |
| 81 | Cxxc1 | 12280 | -0.009 | 0.3023 | No |
| 82 | Dcp1a | 12301 | -0.009 | 0.3017 | No |
| 83 | Atf3 | 12314 | -0.010 | 0.3016 | No |
| 84 | Ddx10 | 12397 | -0.013 | 0.2985 | No |
| 85 | Dkc1 | 12690 | -0.023 | 0.2869 | No |
| 86 | Paip1 | 13091 | -0.037 | 0.2712 | No |
| 87 | Sec11a | 13265 | -0.043 | 0.2653 | No |
| 88 | Vegfa | 13346 | -0.046 | 0.2635 | No |
| 89 | Atf4 | 14362 | -0.081 | 0.2232 | No |
| 90 | Nhp2 | 14381 | -0.082 | 0.2253 | No |
| 91 | Yif1a | 14601 | -0.090 | 0.2191 | No |
| 92 | Exosc5 | 15204 | -0.113 | 0.1974 | No |
| 93 | Srprb | 15964 | -0.141 | 0.1701 | No |
| 94 | Lsm4 | 16712 | -0.169 | 0.1442 | No |
| 95 | Cnot4 | 16769 | -0.172 | 0.1478 | No |
| 96 | Ern1 | 17337 | -0.191 | 0.1303 | No |
| 97 | Cks1b | 17582 | -0.200 | 0.1269 | No |
| 98 | Eef2 | 17619 | -0.202 | 0.1324 | No |
| 99 | Arxes2 | 18319 | -0.230 | 0.1107 | No |
| 100 | H2ax | 18546 | -0.238 | 0.1094 | No |
| 101 | Spcs1 | 19485 | -0.283 | 0.0794 | No |
| 102 | Nfya | 19637 | -0.291 | 0.0831 | No |
| 103 | Wfs1 | 20434 | -0.332 | 0.0608 | No |
| 104 | Banf1 | 21645 | -0.419 | 0.0239 | No |
| 105 | Exosc1 | 22119 | -0.459 | 0.0198 | No |
| 106 | Imp3 | 22877 | -0.556 | 0.0070 | No |
| 107 | Lsm1 | 23391 | -0.711 | 0.0099 | No |