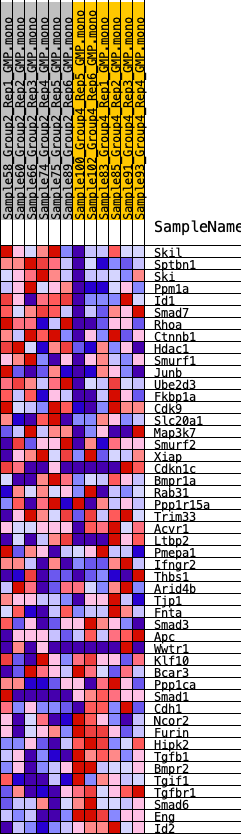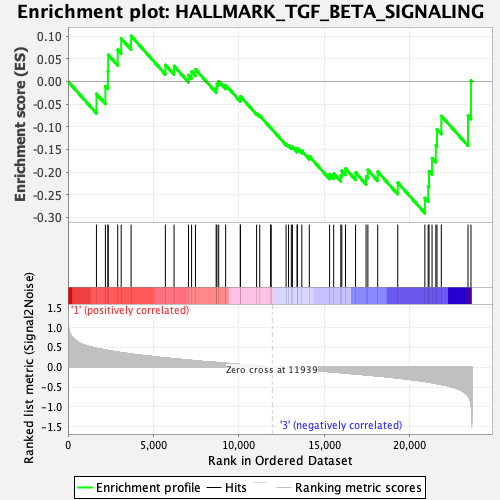
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group2_versus_Group4.GMP.mono_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_TGF_BETA_SIGNALING |
| Enrichment Score (ES) | -0.29049477 |
| Normalized Enrichment Score (NES) | -1.2916238 |
| Nominal p-value | 0.14373717 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.767 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Skil | 1664 | 0.464 | -0.0276 | No |
| 2 | Sptbn1 | 2187 | 0.425 | -0.0105 | No |
| 3 | Ski | 2333 | 0.412 | 0.0214 | No |
| 4 | Ppm1a | 2352 | 0.410 | 0.0586 | No |
| 5 | Id1 | 2908 | 0.376 | 0.0698 | No |
| 6 | Smad7 | 3112 | 0.361 | 0.0946 | No |
| 7 | Rhoa | 3691 | 0.328 | 0.1004 | No |
| 8 | Ctnnb1 | 5696 | 0.228 | 0.0365 | No |
| 9 | Hdac1 | 6204 | 0.205 | 0.0339 | No |
| 10 | Smurf1 | 7051 | 0.172 | 0.0140 | No |
| 11 | Junb | 7230 | 0.166 | 0.0217 | No |
| 12 | Ube2d3 | 7456 | 0.156 | 0.0267 | No |
| 13 | Fkbp1a | 8667 | 0.115 | -0.0140 | No |
| 14 | Cdk9 | 8713 | 0.114 | -0.0054 | No |
| 15 | Slc20a1 | 8817 | 0.110 | 0.0004 | No |
| 16 | Map3k7 | 9223 | 0.094 | -0.0081 | No |
| 17 | Smurf2 | 10077 | 0.063 | -0.0384 | No |
| 18 | Xiap | 10089 | 0.063 | -0.0330 | No |
| 19 | Cdkn1c | 11033 | 0.030 | -0.0702 | No |
| 20 | Bmpr1a | 11217 | 0.025 | -0.0757 | No |
| 21 | Rab31 | 11854 | 0.003 | -0.1024 | No |
| 22 | Ppp1r15a | 11887 | 0.002 | -0.1036 | No |
| 23 | Trim33 | 12758 | -0.025 | -0.1382 | No |
| 24 | Acvr1 | 12899 | -0.030 | -0.1414 | No |
| 25 | Ltbp2 | 13066 | -0.036 | -0.1452 | No |
| 26 | Pmepa1 | 13128 | -0.038 | -0.1443 | No |
| 27 | Ifngr2 | 13402 | -0.048 | -0.1514 | No |
| 28 | Thbs1 | 13420 | -0.049 | -0.1476 | No |
| 29 | Arid4b | 13679 | -0.058 | -0.1533 | No |
| 30 | Tjp1 | 14117 | -0.072 | -0.1652 | No |
| 31 | Fnta | 15304 | -0.117 | -0.2046 | No |
| 32 | Smad3 | 15543 | -0.126 | -0.2031 | No |
| 33 | Apc | 15960 | -0.141 | -0.2077 | No |
| 34 | Wwtr1 | 16018 | -0.143 | -0.1970 | No |
| 35 | Klf10 | 16234 | -0.151 | -0.1921 | No |
| 36 | Bcar3 | 16823 | -0.173 | -0.2011 | No |
| 37 | Ppp1ca | 17438 | -0.195 | -0.2091 | No |
| 38 | Smad1 | 17544 | -0.199 | -0.1951 | No |
| 39 | Cdh1 | 18118 | -0.221 | -0.1990 | No |
| 40 | Ncor2 | 19291 | -0.275 | -0.2233 | No |
| 41 | Furin | 20877 | -0.361 | -0.2571 | Yes |
| 42 | Hipk2 | 21071 | -0.374 | -0.2307 | Yes |
| 43 | Tgfb1 | 21120 | -0.378 | -0.1978 | Yes |
| 44 | Bmpr2 | 21298 | -0.391 | -0.1692 | Yes |
| 45 | Tgif1 | 21519 | -0.408 | -0.1408 | Yes |
| 46 | Tgfbr1 | 21587 | -0.413 | -0.1055 | Yes |
| 47 | Smad6 | 21840 | -0.436 | -0.0759 | Yes |
| 48 | Eng | 23401 | -0.716 | -0.0758 | Yes |
| 49 | Id2 | 23574 | -0.922 | 0.0021 | Yes |