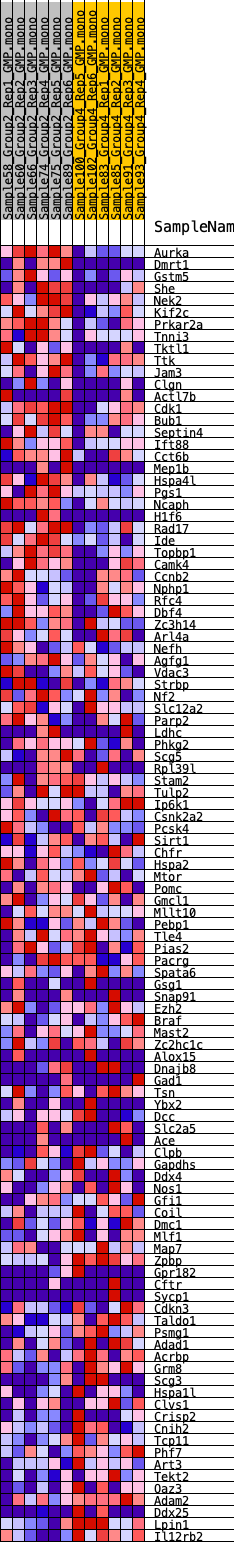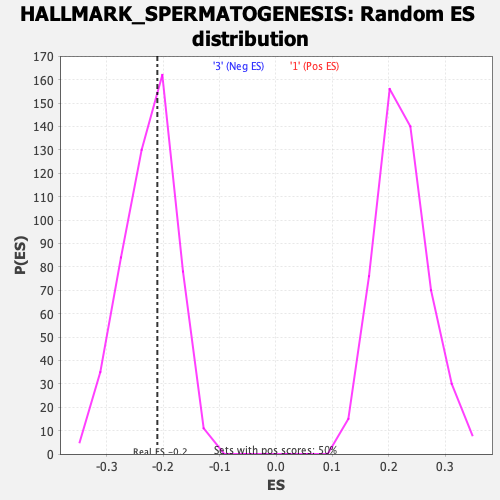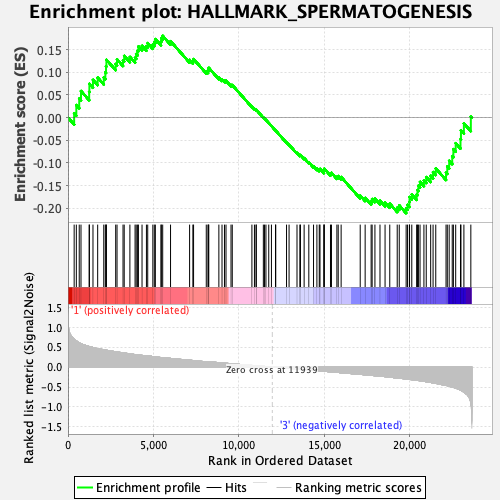
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group2_versus_Group4.GMP.mono_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | -0.21060416 |
| Normalized Enrichment Score (NES) | -0.9368572 |
| Nominal p-value | 0.57227725 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Aurka | 360 | 0.696 | 0.0097 | No |
| 2 | Dmrt1 | 487 | 0.654 | 0.0278 | No |
| 3 | Gstm5 | 657 | 0.603 | 0.0422 | No |
| 4 | She | 760 | 0.583 | 0.0587 | No |
| 5 | Nek2 | 1238 | 0.513 | 0.0568 | No |
| 6 | Kif2c | 1252 | 0.512 | 0.0746 | No |
| 7 | Prkar2a | 1461 | 0.489 | 0.0833 | No |
| 8 | Tnni3 | 1736 | 0.461 | 0.0882 | No |
| 9 | Tktl1 | 2093 | 0.433 | 0.0886 | No |
| 10 | Ttk | 2184 | 0.426 | 0.1000 | No |
| 11 | Jam3 | 2225 | 0.422 | 0.1134 | No |
| 12 | Clgn | 2248 | 0.420 | 0.1276 | No |
| 13 | Actl7b | 2790 | 0.384 | 0.1183 | No |
| 14 | Cdk1 | 2868 | 0.379 | 0.1286 | No |
| 15 | Bub1 | 3219 | 0.355 | 0.1265 | No |
| 16 | Septin4 | 3288 | 0.350 | 0.1361 | No |
| 17 | Ift88 | 3621 | 0.331 | 0.1339 | No |
| 18 | Cct6b | 3924 | 0.310 | 0.1322 | No |
| 19 | Mep1b | 3990 | 0.308 | 0.1405 | No |
| 20 | Hspa4l | 4072 | 0.307 | 0.1480 | No |
| 21 | Pgs1 | 4119 | 0.304 | 0.1569 | No |
| 22 | Ncaph | 4331 | 0.291 | 0.1584 | No |
| 23 | H1f6 | 4588 | 0.281 | 0.1576 | No |
| 24 | Rad17 | 4664 | 0.278 | 0.1644 | No |
| 25 | Ide | 4964 | 0.261 | 0.1610 | No |
| 26 | Topbp1 | 5049 | 0.257 | 0.1667 | No |
| 27 | Camk4 | 5110 | 0.254 | 0.1732 | No |
| 28 | Ccnb2 | 5438 | 0.237 | 0.1678 | No |
| 29 | Nphp1 | 5460 | 0.236 | 0.1754 | No |
| 30 | Rfc4 | 5540 | 0.232 | 0.1803 | No |
| 31 | Dbf4 | 5999 | 0.215 | 0.1686 | No |
| 32 | Zc3h14 | 7114 | 0.170 | 0.1273 | No |
| 33 | Arl4a | 7306 | 0.163 | 0.1250 | No |
| 34 | Nefh | 7346 | 0.161 | 0.1291 | No |
| 35 | Agfg1 | 8089 | 0.131 | 0.1023 | No |
| 36 | Vdac3 | 8190 | 0.127 | 0.1026 | No |
| 37 | Strbp | 8213 | 0.126 | 0.1062 | No |
| 38 | Nf2 | 8236 | 0.125 | 0.1097 | No |
| 39 | Slc12a2 | 8826 | 0.109 | 0.0886 | No |
| 40 | Parp2 | 9010 | 0.103 | 0.0845 | No |
| 41 | Ldhc | 9154 | 0.097 | 0.0819 | No |
| 42 | Phkg2 | 9243 | 0.094 | 0.0815 | No |
| 43 | Scg5 | 9537 | 0.084 | 0.0721 | No |
| 44 | Rpl39l | 9610 | 0.081 | 0.0719 | No |
| 45 | Stam2 | 10759 | 0.040 | 0.0245 | No |
| 46 | Tulp2 | 10907 | 0.035 | 0.0195 | No |
| 47 | Ip6k1 | 11004 | 0.031 | 0.0166 | No |
| 48 | Csnk2a2 | 11013 | 0.031 | 0.0173 | No |
| 49 | Pcsk4 | 11428 | 0.018 | 0.0004 | No |
| 50 | Sirt1 | 11498 | 0.015 | -0.0020 | No |
| 51 | Chfr | 11580 | 0.012 | -0.0051 | No |
| 52 | Hspa2 | 11744 | 0.007 | -0.0118 | No |
| 53 | Mtor | 11901 | 0.001 | -0.0183 | No |
| 54 | Pomc | 12139 | -0.003 | -0.0283 | No |
| 55 | Gmcl1 | 12154 | -0.004 | -0.0288 | No |
| 56 | Mllt10 | 12785 | -0.026 | -0.0546 | No |
| 57 | Pebp1 | 12933 | -0.031 | -0.0598 | No |
| 58 | Tle4 | 13397 | -0.048 | -0.0778 | No |
| 59 | Pias2 | 13564 | -0.054 | -0.0829 | No |
| 60 | Pacrg | 13600 | -0.055 | -0.0824 | No |
| 61 | Spata6 | 13814 | -0.062 | -0.0893 | No |
| 62 | Gsg1 | 14088 | -0.071 | -0.0983 | No |
| 63 | Snap91 | 14364 | -0.081 | -0.1071 | No |
| 64 | Ezh2 | 14561 | -0.089 | -0.1123 | No |
| 65 | Braf | 14699 | -0.094 | -0.1147 | No |
| 66 | Mast2 | 14742 | -0.095 | -0.1131 | No |
| 67 | Zc2hc1c | 14971 | -0.104 | -0.1191 | No |
| 68 | Alox15 | 14980 | -0.104 | -0.1157 | No |
| 69 | Dnajb8 | 14994 | -0.104 | -0.1125 | No |
| 70 | Gad1 | 15356 | -0.119 | -0.1236 | No |
| 71 | Tsn | 15413 | -0.121 | -0.1216 | No |
| 72 | Ybx2 | 15731 | -0.132 | -0.1304 | No |
| 73 | Dcc | 15808 | -0.135 | -0.1288 | No |
| 74 | Slc2a5 | 15979 | -0.141 | -0.1310 | No |
| 75 | Ace | 17095 | -0.184 | -0.1718 | No |
| 76 | Clpb | 17384 | -0.193 | -0.1771 | No |
| 77 | Gapdhs | 17734 | -0.206 | -0.1845 | No |
| 78 | Ddx4 | 17792 | -0.209 | -0.1795 | No |
| 79 | Nos1 | 17956 | -0.216 | -0.1787 | No |
| 80 | Gfi1 | 18252 | -0.226 | -0.1831 | No |
| 81 | Coil | 18549 | -0.239 | -0.1872 | No |
| 82 | Dmc1 | 18824 | -0.251 | -0.1898 | No |
| 83 | Mlf1 | 19260 | -0.274 | -0.1985 | No |
| 84 | Map7 | 19379 | -0.279 | -0.1935 | No |
| 85 | Zpbp | 19782 | -0.299 | -0.1999 | Yes |
| 86 | Gpr182 | 19865 | -0.305 | -0.1924 | Yes |
| 87 | Cftr | 19973 | -0.310 | -0.1859 | Yes |
| 88 | Sycp1 | 19981 | -0.310 | -0.1751 | Yes |
| 89 | Cdkn3 | 20112 | -0.313 | -0.1694 | Yes |
| 90 | Taldo1 | 20397 | -0.330 | -0.1697 | Yes |
| 91 | Psmg1 | 20444 | -0.333 | -0.1597 | Yes |
| 92 | Adad1 | 20500 | -0.336 | -0.1500 | Yes |
| 93 | Acrbp | 20595 | -0.342 | -0.1417 | Yes |
| 94 | Grm8 | 20822 | -0.358 | -0.1385 | Yes |
| 95 | Scg3 | 20958 | -0.367 | -0.1311 | Yes |
| 96 | Hspa1l | 21217 | -0.385 | -0.1283 | Yes |
| 97 | Clvs1 | 21358 | -0.395 | -0.1200 | Yes |
| 98 | Crisp2 | 21515 | -0.407 | -0.1121 | Yes |
| 99 | Cnih2 | 22116 | -0.459 | -0.1211 | Yes |
| 100 | Tcp11 | 22189 | -0.465 | -0.1075 | Yes |
| 101 | Phf7 | 22300 | -0.478 | -0.0951 | Yes |
| 102 | Art3 | 22478 | -0.497 | -0.0848 | Yes |
| 103 | Tekt2 | 22546 | -0.506 | -0.0695 | Yes |
| 104 | Oaz3 | 22681 | -0.525 | -0.0564 | Yes |
| 105 | Adam2 | 22962 | -0.576 | -0.0476 | Yes |
| 106 | Ddx25 | 22989 | -0.583 | -0.0278 | Yes |
| 107 | Lpin1 | 23161 | -0.625 | -0.0127 | Yes |
| 108 | Il12rb2 | 23567 | -0.902 | 0.0024 | Yes |