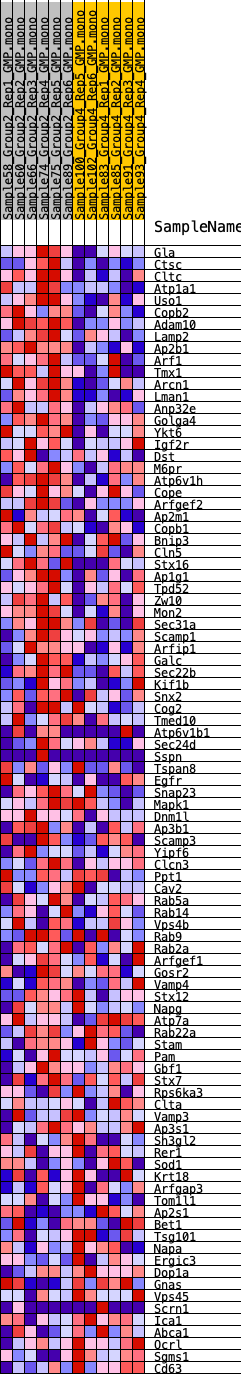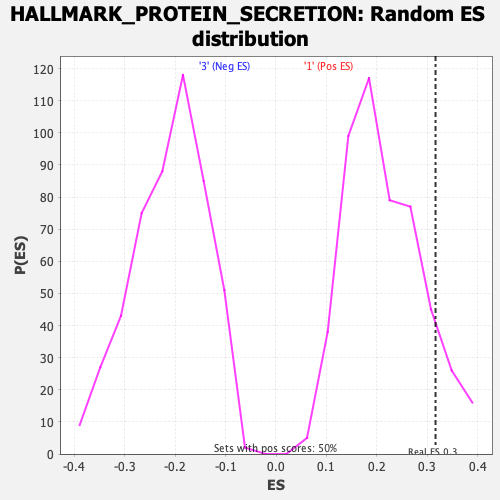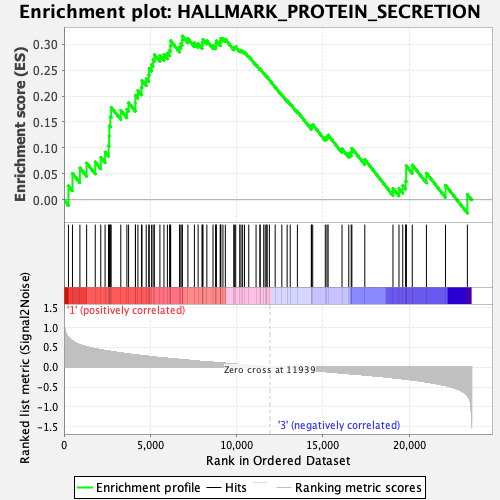
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group2_versus_Group4.GMP.mono_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | 0.315951 |
| Normalized Enrichment Score (NES) | 1.4636317 |
| Nominal p-value | 0.10358566 |
| FDR q-value | 0.24282362 |
| FWER p-Value | 0.487 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gla | 257 | 0.740 | 0.0272 | Yes |
| 2 | Ctsc | 491 | 0.654 | 0.0511 | Yes |
| 3 | Cltc | 922 | 0.558 | 0.0615 | Yes |
| 4 | Atp1a1 | 1312 | 0.503 | 0.0709 | Yes |
| 5 | Uso1 | 1811 | 0.454 | 0.0732 | Yes |
| 6 | Copb2 | 2131 | 0.430 | 0.0818 | Yes |
| 7 | Adam10 | 2381 | 0.409 | 0.0923 | Yes |
| 8 | Lamp2 | 2580 | 0.395 | 0.1042 | Yes |
| 9 | Ap2b1 | 2615 | 0.394 | 0.1231 | Yes |
| 10 | Arf1 | 2632 | 0.392 | 0.1426 | Yes |
| 11 | Tmx1 | 2688 | 0.388 | 0.1603 | Yes |
| 12 | Arcn1 | 2732 | 0.386 | 0.1784 | Yes |
| 13 | Lman1 | 3292 | 0.349 | 0.1727 | Yes |
| 14 | Anp32e | 3644 | 0.329 | 0.1747 | Yes |
| 15 | Golga4 | 3743 | 0.323 | 0.1872 | Yes |
| 16 | Ykt6 | 4138 | 0.303 | 0.1861 | Yes |
| 17 | Igf2r | 4141 | 0.302 | 0.2016 | Yes |
| 18 | Dst | 4284 | 0.294 | 0.2107 | Yes |
| 19 | M6pr | 4491 | 0.286 | 0.2166 | Yes |
| 20 | Atp6v1h | 4520 | 0.284 | 0.2301 | Yes |
| 21 | Cope | 4765 | 0.272 | 0.2337 | Yes |
| 22 | Arfgef2 | 4914 | 0.265 | 0.2411 | Yes |
| 23 | Ap2m1 | 4938 | 0.263 | 0.2536 | Yes |
| 24 | Copb1 | 5077 | 0.256 | 0.2609 | Yes |
| 25 | Bnip3 | 5163 | 0.251 | 0.2703 | Yes |
| 26 | Cln5 | 5237 | 0.247 | 0.2799 | Yes |
| 27 | Stx16 | 5559 | 0.231 | 0.2782 | Yes |
| 28 | Ap1g1 | 5793 | 0.225 | 0.2799 | Yes |
| 29 | Tpd52 | 5991 | 0.215 | 0.2826 | Yes |
| 30 | Zw10 | 6118 | 0.208 | 0.2880 | Yes |
| 31 | Mon2 | 6163 | 0.206 | 0.2968 | Yes |
| 32 | Sec31a | 6182 | 0.206 | 0.3066 | Yes |
| 33 | Scamp1 | 6697 | 0.188 | 0.2944 | Yes |
| 34 | Arfip1 | 6772 | 0.185 | 0.3008 | Yes |
| 35 | Galc | 6847 | 0.182 | 0.3070 | Yes |
| 36 | Sec22b | 6857 | 0.181 | 0.3160 | Yes |
| 37 | Kif1b | 7181 | 0.168 | 0.3109 | No |
| 38 | Snx2 | 7557 | 0.152 | 0.3028 | No |
| 39 | Cog2 | 7773 | 0.143 | 0.3010 | No |
| 40 | Tmed10 | 8006 | 0.133 | 0.2980 | No |
| 41 | Atp6v1b1 | 8015 | 0.133 | 0.3045 | No |
| 42 | Sec24d | 8055 | 0.132 | 0.3097 | No |
| 43 | Sspn | 8279 | 0.124 | 0.3066 | No |
| 44 | Tspan8 | 8635 | 0.117 | 0.2975 | No |
| 45 | Egfr | 8793 | 0.111 | 0.2965 | No |
| 46 | Snap23 | 8805 | 0.110 | 0.3017 | No |
| 47 | Mapk1 | 8827 | 0.109 | 0.3065 | No |
| 48 | Dnm1l | 9058 | 0.101 | 0.3019 | No |
| 49 | Ap3b1 | 9063 | 0.101 | 0.3070 | No |
| 50 | Scamp3 | 9092 | 0.100 | 0.3109 | No |
| 51 | Yipf6 | 9211 | 0.095 | 0.3108 | No |
| 52 | Clcn3 | 9350 | 0.090 | 0.3096 | No |
| 53 | Ppt1 | 9838 | 0.072 | 0.2926 | No |
| 54 | Cav2 | 9890 | 0.070 | 0.2940 | No |
| 55 | Rab5a | 9952 | 0.068 | 0.2949 | No |
| 56 | Rab14 | 10185 | 0.060 | 0.2882 | No |
| 57 | Vps4b | 10260 | 0.057 | 0.2880 | No |
| 58 | Rab9 | 10353 | 0.054 | 0.2868 | No |
| 59 | Rab2a | 10464 | 0.050 | 0.2847 | No |
| 60 | Arfgef1 | 10708 | 0.042 | 0.2766 | No |
| 61 | Gosr2 | 11131 | 0.028 | 0.2601 | No |
| 62 | Vamp4 | 11359 | 0.020 | 0.2515 | No |
| 63 | Stx12 | 11365 | 0.020 | 0.2522 | No |
| 64 | Napg | 11575 | 0.012 | 0.2440 | No |
| 65 | Atp7a | 11662 | 0.009 | 0.2408 | No |
| 66 | Rab22a | 11760 | 0.006 | 0.2370 | No |
| 67 | Stam | 11764 | 0.006 | 0.2371 | No |
| 68 | Pam | 11903 | 0.001 | 0.2314 | No |
| 69 | Gbf1 | 12238 | -0.007 | 0.2175 | No |
| 70 | Stx7 | 12624 | -0.020 | 0.2022 | No |
| 71 | Rps6ka3 | 12932 | -0.031 | 0.1907 | No |
| 72 | Clta | 13111 | -0.037 | 0.1851 | No |
| 73 | Vamp3 | 13526 | -0.052 | 0.1702 | No |
| 74 | Ap3s1 | 14332 | -0.080 | 0.1401 | No |
| 75 | Sh3gl2 | 14346 | -0.080 | 0.1437 | No |
| 76 | Rer1 | 14414 | -0.083 | 0.1451 | No |
| 77 | Sod1 | 15144 | -0.111 | 0.1199 | No |
| 78 | Krt18 | 15240 | -0.114 | 0.1217 | No |
| 79 | Arfgap3 | 15307 | -0.117 | 0.1249 | No |
| 80 | Tom1l1 | 16113 | -0.146 | 0.0982 | No |
| 81 | Ap2s1 | 16503 | -0.160 | 0.0899 | No |
| 82 | Bet1 | 16644 | -0.167 | 0.0926 | No |
| 83 | Tsg101 | 16684 | -0.169 | 0.0996 | No |
| 84 | Napa | 17433 | -0.195 | 0.0779 | No |
| 85 | Ergic3 | 19066 | -0.264 | 0.0221 | No |
| 86 | Dop1a | 19414 | -0.279 | 0.0218 | No |
| 87 | Gnas | 19633 | -0.290 | 0.0275 | No |
| 88 | Vps45 | 19801 | -0.301 | 0.0359 | No |
| 89 | Scrn1 | 19831 | -0.303 | 0.0502 | No |
| 90 | Ica1 | 19832 | -0.303 | 0.0658 | No |
| 91 | Abca1 | 20184 | -0.318 | 0.0673 | No |
| 92 | Ocrl | 21005 | -0.370 | 0.0515 | No |
| 93 | Sgms1 | 22111 | -0.458 | 0.0282 | No |
| 94 | Cd63 | 23377 | -0.700 | 0.0105 | No |