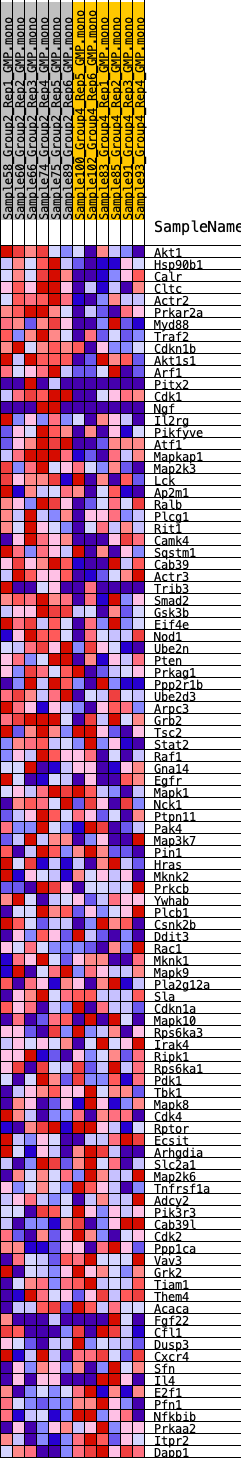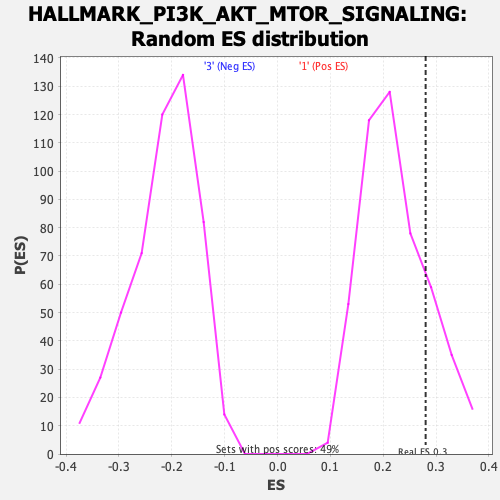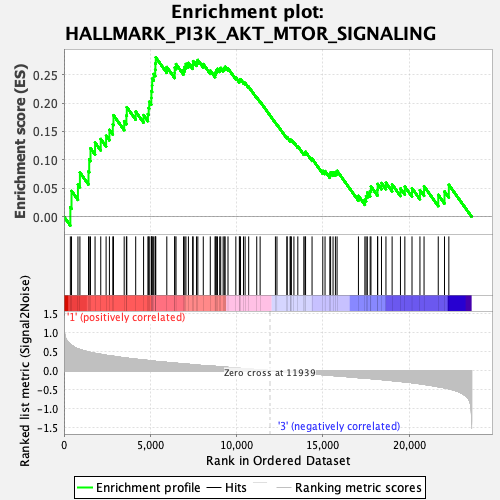
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group2_versus_Group4.GMP.mono_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | 0.2802652 |
| Normalized Enrichment Score (NES) | 1.2587402 |
| Nominal p-value | 0.20162933 |
| FDR q-value | 0.5875996 |
| FWER p-Value | 0.812 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Akt1 | 368 | 0.690 | 0.0167 | Yes |
| 2 | Hsp90b1 | 436 | 0.671 | 0.0453 | Yes |
| 3 | Calr | 808 | 0.574 | 0.0564 | Yes |
| 4 | Cltc | 922 | 0.558 | 0.0778 | Yes |
| 5 | Actr2 | 1420 | 0.493 | 0.0798 | Yes |
| 6 | Prkar2a | 1461 | 0.489 | 0.1010 | Yes |
| 7 | Myd88 | 1536 | 0.480 | 0.1204 | Yes |
| 8 | Traf2 | 1800 | 0.455 | 0.1305 | Yes |
| 9 | Cdkn1b | 2130 | 0.430 | 0.1367 | Yes |
| 10 | Akt1s1 | 2440 | 0.405 | 0.1426 | Yes |
| 11 | Arf1 | 2632 | 0.392 | 0.1528 | Yes |
| 12 | Pitx2 | 2829 | 0.382 | 0.1624 | Yes |
| 13 | Cdk1 | 2868 | 0.379 | 0.1786 | Yes |
| 14 | Ngf | 3488 | 0.338 | 0.1681 | Yes |
| 15 | Il2rg | 3616 | 0.331 | 0.1782 | Yes |
| 16 | Pikfyve | 3640 | 0.329 | 0.1926 | Yes |
| 17 | Atf1 | 4155 | 0.301 | 0.1849 | Yes |
| 18 | Mapkap1 | 4612 | 0.281 | 0.1787 | Yes |
| 19 | Map2k3 | 4864 | 0.267 | 0.1806 | Yes |
| 20 | Lck | 4909 | 0.265 | 0.1911 | Yes |
| 21 | Ap2m1 | 4938 | 0.263 | 0.2023 | Yes |
| 22 | Ralb | 5059 | 0.256 | 0.2092 | Yes |
| 23 | Plcg1 | 5065 | 0.256 | 0.2210 | Yes |
| 24 | Rit1 | 5104 | 0.254 | 0.2313 | Yes |
| 25 | Camk4 | 5110 | 0.254 | 0.2430 | Yes |
| 26 | Sqstm1 | 5196 | 0.249 | 0.2510 | Yes |
| 27 | Cab39 | 5285 | 0.245 | 0.2588 | Yes |
| 28 | Actr3 | 5297 | 0.244 | 0.2698 | Yes |
| 29 | Trib3 | 5319 | 0.243 | 0.2803 | Yes |
| 30 | Smad2 | 5958 | 0.217 | 0.2633 | No |
| 31 | Gsk3b | 6415 | 0.198 | 0.2532 | No |
| 32 | Eif4e | 6419 | 0.198 | 0.2624 | No |
| 33 | Nod1 | 6492 | 0.195 | 0.2685 | No |
| 34 | Ube2n | 6927 | 0.178 | 0.2584 | No |
| 35 | Pten | 6998 | 0.175 | 0.2636 | No |
| 36 | Prkag1 | 7066 | 0.172 | 0.2688 | No |
| 37 | Ppp2r1b | 7211 | 0.167 | 0.2705 | No |
| 38 | Ube2d3 | 7456 | 0.156 | 0.2675 | No |
| 39 | Arpc3 | 7483 | 0.155 | 0.2736 | No |
| 40 | Grb2 | 7678 | 0.147 | 0.2723 | No |
| 41 | Tsc2 | 7756 | 0.143 | 0.2757 | No |
| 42 | Stat2 | 8072 | 0.132 | 0.2685 | No |
| 43 | Raf1 | 8478 | 0.123 | 0.2571 | No |
| 44 | Gna14 | 8753 | 0.112 | 0.2507 | No |
| 45 | Egfr | 8793 | 0.111 | 0.2542 | No |
| 46 | Mapk1 | 8827 | 0.109 | 0.2579 | No |
| 47 | Nck1 | 8894 | 0.107 | 0.2601 | No |
| 48 | Ptpn11 | 9024 | 0.102 | 0.2595 | No |
| 49 | Pak4 | 9082 | 0.100 | 0.2617 | No |
| 50 | Map3k7 | 9223 | 0.094 | 0.2602 | No |
| 51 | Pin1 | 9289 | 0.092 | 0.2618 | No |
| 52 | Hras | 9340 | 0.090 | 0.2639 | No |
| 53 | Mknk2 | 9505 | 0.085 | 0.2609 | No |
| 54 | Prkcb | 9959 | 0.068 | 0.2448 | No |
| 55 | Ywhab | 10163 | 0.061 | 0.2390 | No |
| 56 | Plcb1 | 10182 | 0.060 | 0.2411 | No |
| 57 | Csnk2b | 10229 | 0.058 | 0.2419 | No |
| 58 | Ddit3 | 10402 | 0.052 | 0.2370 | No |
| 59 | Rac1 | 10496 | 0.049 | 0.2353 | No |
| 60 | Mknk1 | 10703 | 0.042 | 0.2285 | No |
| 61 | Mapk9 | 11170 | 0.027 | 0.2100 | No |
| 62 | Pla2g12a | 11367 | 0.019 | 0.2025 | No |
| 63 | Sla | 12253 | -0.007 | 0.1653 | No |
| 64 | Cdkn1a | 12342 | -0.011 | 0.1620 | No |
| 65 | Mapk10 | 12918 | -0.030 | 0.1390 | No |
| 66 | Rps6ka3 | 12932 | -0.031 | 0.1399 | No |
| 67 | Irak4 | 13109 | -0.037 | 0.1342 | No |
| 68 | Ripk1 | 13119 | -0.037 | 0.1355 | No |
| 69 | Rps6ka1 | 13179 | -0.040 | 0.1349 | No |
| 70 | Pdk1 | 13325 | -0.045 | 0.1308 | No |
| 71 | Tbk1 | 13551 | -0.053 | 0.1238 | No |
| 72 | Mapk8 | 13899 | -0.064 | 0.1120 | No |
| 73 | Cdk4 | 13966 | -0.067 | 0.1124 | No |
| 74 | Rptor | 14000 | -0.068 | 0.1142 | No |
| 75 | Ecsit | 14376 | -0.082 | 0.1021 | No |
| 76 | Arhgdia | 14998 | -0.105 | 0.0806 | No |
| 77 | Slc2a1 | 15129 | -0.110 | 0.0802 | No |
| 78 | Map2k6 | 15407 | -0.120 | 0.0741 | No |
| 79 | Tnfrsf1a | 15451 | -0.122 | 0.0780 | No |
| 80 | Adcy2 | 15593 | -0.128 | 0.0780 | No |
| 81 | Pik3r3 | 15730 | -0.132 | 0.0784 | No |
| 82 | Cab39l | 15826 | -0.136 | 0.0807 | No |
| 83 | Cdk2 | 17064 | -0.183 | 0.0367 | No |
| 84 | Ppp1ca | 17438 | -0.195 | 0.0300 | No |
| 85 | Vav3 | 17508 | -0.197 | 0.0363 | No |
| 86 | Grk2 | 17580 | -0.200 | 0.0427 | No |
| 87 | Tiam1 | 17744 | -0.206 | 0.0454 | No |
| 88 | Them4 | 17790 | -0.208 | 0.0533 | No |
| 89 | Acaca | 18171 | -0.222 | 0.0475 | No |
| 90 | Fgf22 | 18181 | -0.223 | 0.0576 | No |
| 91 | Cfl1 | 18401 | -0.233 | 0.0592 | No |
| 92 | Dusp3 | 18659 | -0.244 | 0.0597 | No |
| 93 | Cxcr4 | 19016 | -0.261 | 0.0569 | No |
| 94 | Sfn | 19500 | -0.284 | 0.0496 | No |
| 95 | Il4 | 19754 | -0.298 | 0.0528 | No |
| 96 | E2f1 | 20169 | -0.317 | 0.0501 | No |
| 97 | Pfn1 | 20629 | -0.345 | 0.0467 | No |
| 98 | Nfkbib | 20867 | -0.361 | 0.0536 | No |
| 99 | Prkaa2 | 21690 | -0.423 | 0.0385 | No |
| 100 | Itpr2 | 22052 | -0.453 | 0.0443 | No |
| 101 | Dapp1 | 22304 | -0.479 | 0.0561 | No |