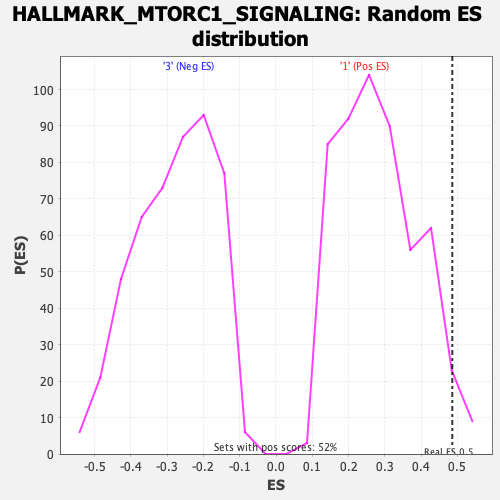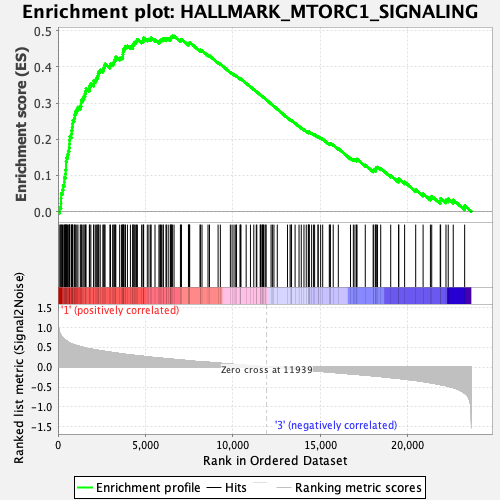
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group2_versus_Group4.GMP.mono_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_MTORC1_SIGNALING |
| Enrichment Score (ES) | 0.48589447 |
| Normalized Enrichment Score (NES) | 1.7037923 |
| Nominal p-value | 0.030534351 |
| FDR q-value | 0.13330366 |
| FWER p-Value | 0.159 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cyp51 | 104 | 0.843 | 0.0108 | Yes |
| 2 | Vldlr | 168 | 0.790 | 0.0224 | Yes |
| 3 | Hspa9 | 174 | 0.786 | 0.0363 | Yes |
| 4 | Hspd1 | 180 | 0.781 | 0.0502 | Yes |
| 5 | Gla | 257 | 0.740 | 0.0604 | Yes |
| 6 | Sytl2 | 291 | 0.728 | 0.0721 | Yes |
| 7 | Aurka | 360 | 0.696 | 0.0817 | Yes |
| 8 | Canx | 363 | 0.694 | 0.0942 | Yes |
| 9 | Hspa4 | 425 | 0.673 | 0.1037 | Yes |
| 10 | Hsp90b1 | 436 | 0.671 | 0.1154 | Yes |
| 11 | Asns | 459 | 0.660 | 0.1264 | Yes |
| 12 | Rpn1 | 460 | 0.660 | 0.1383 | Yes |
| 13 | Ctsc | 491 | 0.654 | 0.1489 | Yes |
| 14 | Etf1 | 547 | 0.634 | 0.1579 | Yes |
| 15 | Hmbs | 583 | 0.622 | 0.1677 | Yes |
| 16 | Rrp9 | 637 | 0.608 | 0.1764 | Yes |
| 17 | Plk1 | 653 | 0.603 | 0.1866 | Yes |
| 18 | Ufm1 | 658 | 0.603 | 0.1974 | Yes |
| 19 | Bhlhe40 | 675 | 0.600 | 0.2075 | Yes |
| 20 | Hmgcr | 766 | 0.581 | 0.2142 | Yes |
| 21 | Ero1a | 776 | 0.580 | 0.2242 | Yes |
| 22 | Calr | 808 | 0.574 | 0.2333 | Yes |
| 23 | Slc7a11 | 829 | 0.569 | 0.2427 | Yes |
| 24 | Hspa5 | 845 | 0.567 | 0.2523 | Yes |
| 25 | Ccnf | 930 | 0.557 | 0.2588 | Yes |
| 26 | Rdh11 | 960 | 0.552 | 0.2675 | Yes |
| 27 | Rrm2 | 988 | 0.548 | 0.2762 | Yes |
| 28 | Slc1a4 | 1073 | 0.535 | 0.2823 | Yes |
| 29 | Stard4 | 1139 | 0.524 | 0.2890 | Yes |
| 30 | Ssr1 | 1273 | 0.510 | 0.2925 | Yes |
| 31 | Edem1 | 1325 | 0.502 | 0.2994 | Yes |
| 32 | Stip1 | 1328 | 0.501 | 0.3084 | Yes |
| 33 | Actr2 | 1420 | 0.493 | 0.3134 | Yes |
| 34 | Hmgcs1 | 1484 | 0.486 | 0.3195 | Yes |
| 35 | Gpi1 | 1544 | 0.479 | 0.3256 | Yes |
| 36 | Serpinh1 | 1576 | 0.474 | 0.3328 | Yes |
| 37 | Ddx39a | 1601 | 0.471 | 0.3403 | Yes |
| 38 | Xbp1 | 1792 | 0.456 | 0.3404 | Yes |
| 39 | Uso1 | 1811 | 0.454 | 0.3479 | Yes |
| 40 | Got1 | 1877 | 0.450 | 0.3532 | Yes |
| 41 | Sqle | 2040 | 0.437 | 0.3542 | Yes |
| 42 | Mthfd2 | 2045 | 0.437 | 0.3619 | Yes |
| 43 | Mcm4 | 2155 | 0.428 | 0.3650 | Yes |
| 44 | Ldlr | 2213 | 0.423 | 0.3702 | Yes |
| 45 | Pno1 | 2274 | 0.418 | 0.3752 | Yes |
| 46 | Nupr1 | 2293 | 0.416 | 0.3819 | Yes |
| 47 | Acsl3 | 2332 | 0.412 | 0.3878 | Yes |
| 48 | Idi1 | 2413 | 0.408 | 0.3917 | Yes |
| 49 | Cyb5b | 2564 | 0.396 | 0.3925 | Yes |
| 50 | Psat1 | 2616 | 0.394 | 0.3974 | Yes |
| 51 | Tm7sf2 | 2642 | 0.392 | 0.4034 | Yes |
| 52 | Cct6a | 2690 | 0.388 | 0.4084 | Yes |
| 53 | Abcf2 | 2968 | 0.369 | 0.4032 | Yes |
| 54 | Gga2 | 3001 | 0.368 | 0.4085 | Yes |
| 55 | Rab1a | 3124 | 0.360 | 0.4098 | Yes |
| 56 | Pfkl | 3192 | 0.356 | 0.4134 | Yes |
| 57 | Bub1 | 3219 | 0.355 | 0.4187 | Yes |
| 58 | Acly | 3277 | 0.351 | 0.4226 | Yes |
| 59 | Atp2a2 | 3304 | 0.348 | 0.4278 | Yes |
| 60 | Eno1b | 3525 | 0.337 | 0.4244 | Yes |
| 61 | Aldoa | 3625 | 0.330 | 0.4262 | Yes |
| 62 | Dhcr24 | 3706 | 0.326 | 0.4287 | Yes |
| 63 | Psme3 | 3708 | 0.326 | 0.4345 | Yes |
| 64 | Itgb2 | 3718 | 0.326 | 0.4400 | Yes |
| 65 | Shmt2 | 3727 | 0.325 | 0.4455 | Yes |
| 66 | Cd9 | 3760 | 0.322 | 0.4500 | Yes |
| 67 | Cth | 3830 | 0.317 | 0.4527 | Yes |
| 68 | Tfrc | 3855 | 0.315 | 0.4574 | Yes |
| 69 | Ppia | 3968 | 0.309 | 0.4582 | Yes |
| 70 | Ykt6 | 4138 | 0.303 | 0.4565 | Yes |
| 71 | Ifi30 | 4275 | 0.294 | 0.4560 | Yes |
| 72 | Nup205 | 4277 | 0.294 | 0.4613 | Yes |
| 73 | Bcat1 | 4343 | 0.290 | 0.4637 | Yes |
| 74 | Psmc4 | 4353 | 0.290 | 0.4686 | Yes |
| 75 | Sc5d | 4444 | 0.289 | 0.4699 | Yes |
| 76 | M6pr | 4491 | 0.286 | 0.4731 | Yes |
| 77 | Serp1 | 4536 | 0.282 | 0.4764 | Yes |
| 78 | Txnrd1 | 4780 | 0.271 | 0.4709 | Yes |
| 79 | Map2k3 | 4864 | 0.267 | 0.4722 | Yes |
| 80 | Gsr | 4876 | 0.267 | 0.4765 | Yes |
| 81 | Hk2 | 4896 | 0.266 | 0.4805 | Yes |
| 82 | Rit1 | 5104 | 0.254 | 0.4763 | Yes |
| 83 | Sqstm1 | 5196 | 0.249 | 0.4769 | Yes |
| 84 | Actr3 | 5297 | 0.244 | 0.4770 | Yes |
| 85 | Trib3 | 5319 | 0.243 | 0.4805 | Yes |
| 86 | Insig1 | 5544 | 0.231 | 0.4751 | Yes |
| 87 | Gtf2h1 | 5770 | 0.227 | 0.4696 | Yes |
| 88 | Pitpnb | 5827 | 0.224 | 0.4713 | Yes |
| 89 | Ifrd1 | 5855 | 0.222 | 0.4741 | Yes |
| 90 | Fdxr | 5910 | 0.219 | 0.4758 | Yes |
| 91 | Fgl2 | 6004 | 0.215 | 0.4757 | Yes |
| 92 | Add3 | 6019 | 0.214 | 0.4789 | Yes |
| 93 | Nampt | 6179 | 0.206 | 0.4759 | Yes |
| 94 | Nufip1 | 6188 | 0.205 | 0.4792 | Yes |
| 95 | Gmps | 6299 | 0.202 | 0.4782 | Yes |
| 96 | Gsk3b | 6415 | 0.198 | 0.4769 | Yes |
| 97 | Psmc6 | 6450 | 0.197 | 0.4790 | Yes |
| 98 | Tmem97 | 6453 | 0.197 | 0.4824 | Yes |
| 99 | Plod2 | 6494 | 0.195 | 0.4842 | Yes |
| 100 | Idh1 | 6538 | 0.193 | 0.4859 | Yes |
| 101 | Slc7a5 | 6632 | 0.189 | 0.4853 | No |
| 102 | Pgk1 | 6996 | 0.175 | 0.4730 | No |
| 103 | Mcm2 | 7029 | 0.174 | 0.4748 | No |
| 104 | Immt | 7055 | 0.172 | 0.4768 | No |
| 105 | Ube2d3 | 7456 | 0.156 | 0.4626 | No |
| 106 | Prdx1 | 7460 | 0.156 | 0.4653 | No |
| 107 | Skap2 | 7514 | 0.154 | 0.4658 | No |
| 108 | Lgmn | 7527 | 0.153 | 0.4680 | No |
| 109 | Psmd14 | 8124 | 0.130 | 0.4449 | No |
| 110 | Cacybp | 8133 | 0.129 | 0.4469 | No |
| 111 | Ccng1 | 8230 | 0.125 | 0.4451 | No |
| 112 | Slc2a3 | 8574 | 0.119 | 0.4326 | No |
| 113 | P4ha1 | 8657 | 0.116 | 0.4312 | No |
| 114 | Ldha | 9150 | 0.098 | 0.4120 | No |
| 115 | Elovl5 | 9279 | 0.093 | 0.4082 | No |
| 116 | Ddit4 | 9851 | 0.071 | 0.3851 | No |
| 117 | Wars1 | 9949 | 0.068 | 0.3822 | No |
| 118 | Pgm1 | 10060 | 0.064 | 0.3787 | No |
| 119 | Fads1 | 10158 | 0.061 | 0.3756 | No |
| 120 | Nfyc | 10204 | 0.059 | 0.3748 | No |
| 121 | Ddit3 | 10402 | 0.052 | 0.3673 | No |
| 122 | Srd5a1 | 10435 | 0.051 | 0.3668 | No |
| 123 | Tpi1 | 10454 | 0.050 | 0.3670 | No |
| 124 | G6pdx | 10754 | 0.040 | 0.3549 | No |
| 125 | Me1 | 10999 | 0.032 | 0.3451 | No |
| 126 | Nfil3 | 11191 | 0.026 | 0.3374 | No |
| 127 | Ebp | 11332 | 0.021 | 0.3318 | No |
| 128 | Psmd13 | 11362 | 0.020 | 0.3309 | No |
| 129 | Sord | 11557 | 0.013 | 0.3229 | No |
| 130 | Tes | 11572 | 0.012 | 0.3225 | No |
| 131 | Psmc2 | 11612 | 0.011 | 0.3210 | No |
| 132 | Psmd12 | 11688 | 0.008 | 0.3180 | No |
| 133 | Slc6a6 | 11695 | 0.008 | 0.3179 | No |
| 134 | Psma4 | 11745 | 0.007 | 0.3159 | No |
| 135 | Cdc25a | 11774 | 0.005 | 0.3148 | No |
| 136 | Ak4 | 11879 | 0.002 | 0.3104 | No |
| 137 | Ppp1r15a | 11887 | 0.002 | 0.3101 | No |
| 138 | Cops5 | 12177 | -0.004 | 0.2979 | No |
| 139 | Sla | 12253 | -0.007 | 0.2948 | No |
| 140 | Cdkn1a | 12342 | -0.011 | 0.2912 | No |
| 141 | Lta4h | 12533 | -0.018 | 0.2835 | No |
| 142 | Tcea1 | 13116 | -0.037 | 0.2593 | No |
| 143 | Sec11a | 13265 | -0.043 | 0.2538 | No |
| 144 | Pdk1 | 13325 | -0.045 | 0.2520 | No |
| 145 | Ung | 13348 | -0.046 | 0.2519 | No |
| 146 | Tbk1 | 13551 | -0.053 | 0.2443 | No |
| 147 | Hspe1 | 13776 | -0.060 | 0.2358 | No |
| 148 | Coro1a | 13904 | -0.065 | 0.2316 | No |
| 149 | Uchl5 | 14058 | -0.069 | 0.2263 | No |
| 150 | Psma3 | 14189 | -0.075 | 0.2221 | No |
| 151 | Nmt1 | 14305 | -0.079 | 0.2186 | No |
| 152 | Ppa1 | 14330 | -0.080 | 0.2190 | No |
| 153 | Glrx | 14350 | -0.081 | 0.2197 | No |
| 154 | Rpa1 | 14352 | -0.081 | 0.2211 | No |
| 155 | Fkbp2 | 14497 | -0.087 | 0.2165 | No |
| 156 | Pdap1 | 14608 | -0.091 | 0.2134 | No |
| 157 | Sdf2l1 | 14667 | -0.092 | 0.2126 | No |
| 158 | Btg2 | 14863 | -0.099 | 0.2061 | No |
| 159 | Gclc | 14866 | -0.100 | 0.2078 | No |
| 160 | Adipor2 | 15011 | -0.105 | 0.2036 | No |
| 161 | Slc2a1 | 15129 | -0.110 | 0.2006 | No |
| 162 | Niban1 | 15506 | -0.124 | 0.1868 | No |
| 163 | Qdpr | 15547 | -0.126 | 0.1873 | No |
| 164 | Phgdh | 15567 | -0.127 | 0.1888 | No |
| 165 | Pik3r3 | 15730 | -0.132 | 0.1843 | No |
| 166 | Tomm40 | 16021 | -0.143 | 0.1745 | No |
| 167 | Tubg1 | 16717 | -0.170 | 0.1479 | No |
| 168 | Psmb5 | 16881 | -0.176 | 0.1441 | No |
| 169 | Elovl6 | 16973 | -0.180 | 0.1435 | No |
| 170 | Gbe1 | 17065 | -0.183 | 0.1429 | No |
| 171 | Dhcr7 | 17088 | -0.184 | 0.1453 | No |
| 172 | Eif2s2 | 17562 | -0.199 | 0.1287 | No |
| 173 | Slc1a5 | 18012 | -0.217 | 0.1134 | No |
| 174 | Igfbp5 | 18049 | -0.219 | 0.1159 | No |
| 175 | Cfp | 18160 | -0.222 | 0.1152 | No |
| 176 | Acaca | 18171 | -0.222 | 0.1188 | No |
| 177 | Tuba4a | 18182 | -0.223 | 0.1224 | No |
| 178 | Fads2 | 18263 | -0.227 | 0.1230 | No |
| 179 | Eef1e1 | 18444 | -0.236 | 0.1196 | No |
| 180 | Cxcr4 | 19016 | -0.261 | 0.1000 | No |
| 181 | Polr3g | 19465 | -0.282 | 0.0859 | No |
| 182 | Dhfr | 19478 | -0.282 | 0.0905 | No |
| 183 | Arpc5l | 19811 | -0.302 | 0.0818 | No |
| 184 | Psmg1 | 20444 | -0.333 | 0.0608 | No |
| 185 | Nfkbib | 20867 | -0.361 | 0.0493 | No |
| 186 | Atp6v1d | 21285 | -0.390 | 0.0386 | No |
| 187 | Mthfd2l | 21360 | -0.396 | 0.0425 | No |
| 188 | Egln3 | 21846 | -0.436 | 0.0297 | No |
| 189 | Mllt11 | 21869 | -0.438 | 0.0367 | No |
| 190 | Slc37a4 | 22175 | -0.463 | 0.0320 | No |
| 191 | Dapp1 | 22304 | -0.479 | 0.0352 | No |
| 192 | Psph | 22590 | -0.513 | 0.0323 | No |
| 193 | Pnp | 23242 | -0.651 | 0.0163 | No |