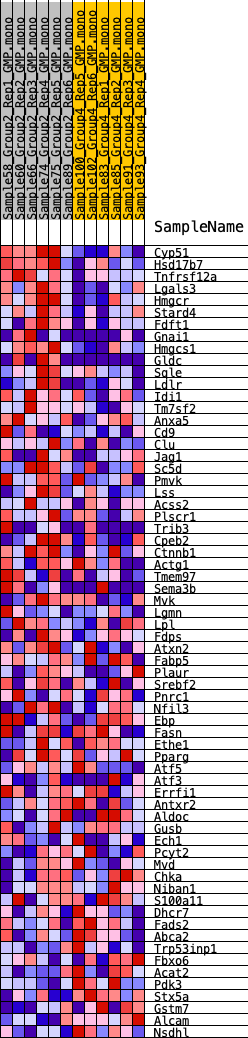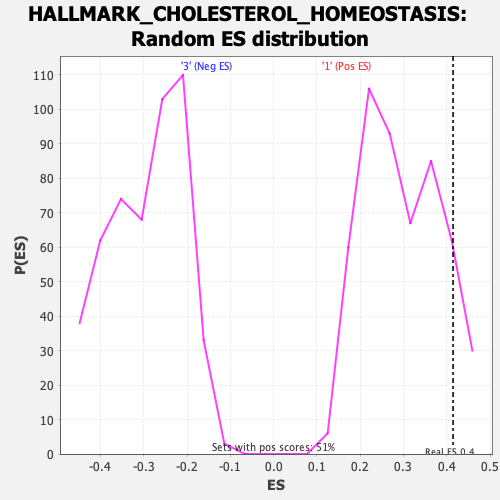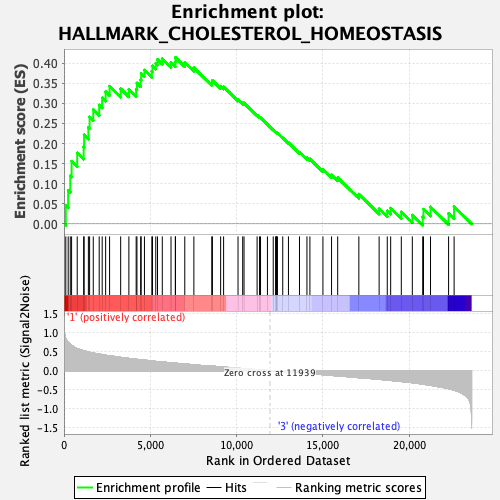
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group2_versus_Group4.GMP.mono_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | 0.4145629 |
| Normalized Enrichment Score (NES) | 1.3980181 |
| Nominal p-value | 0.106090374 |
| FDR q-value | 0.31837273 |
| FWER p-Value | 0.597 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cyp51 | 104 | 0.843 | 0.0455 | Yes |
| 2 | Hsd17b7 | 243 | 0.744 | 0.0837 | Yes |
| 3 | Tnfrsf12a | 364 | 0.694 | 0.1196 | Yes |
| 4 | Lgals3 | 440 | 0.669 | 0.1560 | Yes |
| 5 | Hmgcr | 766 | 0.581 | 0.1766 | Yes |
| 6 | Stard4 | 1139 | 0.524 | 0.1919 | Yes |
| 7 | Fdft1 | 1167 | 0.522 | 0.2216 | Yes |
| 8 | Gnai1 | 1413 | 0.494 | 0.2404 | Yes |
| 9 | Hmgcs1 | 1484 | 0.486 | 0.2662 | Yes |
| 10 | Gldc | 1696 | 0.463 | 0.2846 | Yes |
| 11 | Sqle | 2040 | 0.437 | 0.2960 | Yes |
| 12 | Ldlr | 2213 | 0.423 | 0.3137 | Yes |
| 13 | Idi1 | 2413 | 0.408 | 0.3294 | Yes |
| 14 | Tm7sf2 | 2642 | 0.392 | 0.3429 | Yes |
| 15 | Anxa5 | 3284 | 0.350 | 0.3364 | Yes |
| 16 | Cd9 | 3760 | 0.322 | 0.3353 | Yes |
| 17 | Clu | 4182 | 0.300 | 0.3351 | Yes |
| 18 | Jag1 | 4227 | 0.297 | 0.3509 | Yes |
| 19 | Sc5d | 4444 | 0.289 | 0.3588 | Yes |
| 20 | Pmvk | 4471 | 0.287 | 0.3747 | Yes |
| 21 | Lss | 4662 | 0.278 | 0.3830 | Yes |
| 22 | Acss2 | 5091 | 0.255 | 0.3800 | Yes |
| 23 | Plscr1 | 5129 | 0.253 | 0.3934 | Yes |
| 24 | Trib3 | 5319 | 0.243 | 0.3997 | Yes |
| 25 | Cpeb2 | 5420 | 0.238 | 0.4096 | Yes |
| 26 | Ctnnb1 | 5696 | 0.228 | 0.4114 | Yes |
| 27 | Actg1 | 6202 | 0.205 | 0.4021 | Yes |
| 28 | Tmem97 | 6453 | 0.197 | 0.4031 | Yes |
| 29 | Sema3b | 6459 | 0.197 | 0.4146 | Yes |
| 30 | Mvk | 7000 | 0.175 | 0.4020 | No |
| 31 | Lgmn | 7527 | 0.153 | 0.3887 | No |
| 32 | Lpl | 8564 | 0.119 | 0.3518 | No |
| 33 | Fdps | 8602 | 0.118 | 0.3572 | No |
| 34 | Atxn2 | 9078 | 0.100 | 0.3430 | No |
| 35 | Fabp5 | 9250 | 0.094 | 0.3413 | No |
| 36 | Plaur | 10086 | 0.063 | 0.3096 | No |
| 37 | Srebf2 | 10356 | 0.054 | 0.3013 | No |
| 38 | Pnrc1 | 10433 | 0.051 | 0.3011 | No |
| 39 | Nfil3 | 11191 | 0.026 | 0.2705 | No |
| 40 | Ebp | 11332 | 0.021 | 0.2658 | No |
| 41 | Fasn | 11390 | 0.019 | 0.2645 | No |
| 42 | Ethe1 | 11790 | 0.005 | 0.2479 | No |
| 43 | Pparg | 12126 | -0.003 | 0.2338 | No |
| 44 | Atf5 | 12261 | -0.008 | 0.2286 | No |
| 45 | Atf3 | 12314 | -0.010 | 0.2270 | No |
| 46 | Errfi1 | 12315 | -0.010 | 0.2276 | No |
| 47 | Antxr2 | 12371 | -0.012 | 0.2259 | No |
| 48 | Aldoc | 12677 | -0.022 | 0.2143 | No |
| 49 | Gusb | 13008 | -0.034 | 0.2023 | No |
| 50 | Ech1 | 13648 | -0.057 | 0.1785 | No |
| 51 | Pcyt2 | 14080 | -0.070 | 0.1644 | No |
| 52 | Mvd | 14252 | -0.077 | 0.1617 | No |
| 53 | Chka | 15006 | -0.105 | 0.1359 | No |
| 54 | Niban1 | 15506 | -0.124 | 0.1221 | No |
| 55 | S100a11 | 15862 | -0.137 | 0.1151 | No |
| 56 | Dhcr7 | 17088 | -0.184 | 0.0740 | No |
| 57 | Fads2 | 18263 | -0.227 | 0.0377 | No |
| 58 | Abca2 | 18735 | -0.248 | 0.0323 | No |
| 59 | Trp53inp1 | 18932 | -0.257 | 0.0392 | No |
| 60 | Fbxo6 | 19549 | -0.285 | 0.0299 | No |
| 61 | Acat2 | 20187 | -0.318 | 0.0217 | No |
| 62 | Pdk3 | 20789 | -0.356 | 0.0172 | No |
| 63 | Stx5a | 20835 | -0.358 | 0.0365 | No |
| 64 | Gstm7 | 21242 | -0.387 | 0.0422 | No |
| 65 | Alcam | 22289 | -0.477 | 0.0260 | No |
| 66 | Nsdhl | 22609 | -0.517 | 0.0431 | No |