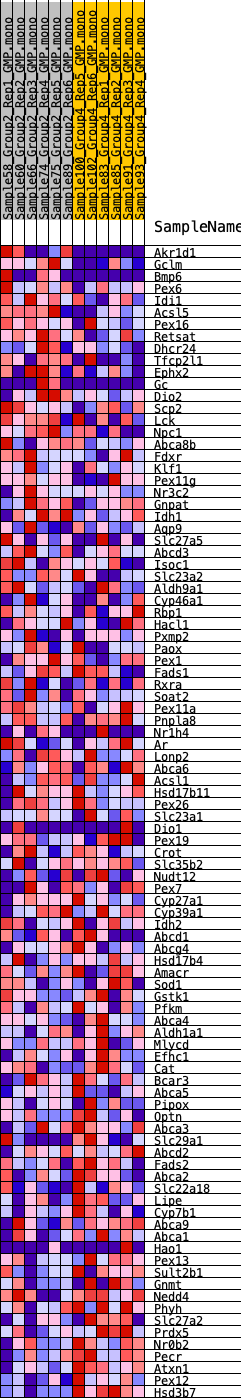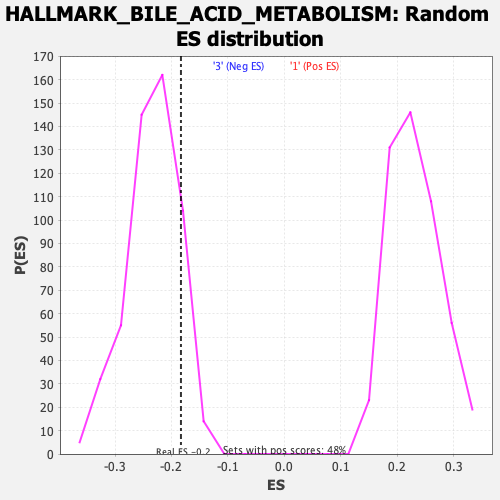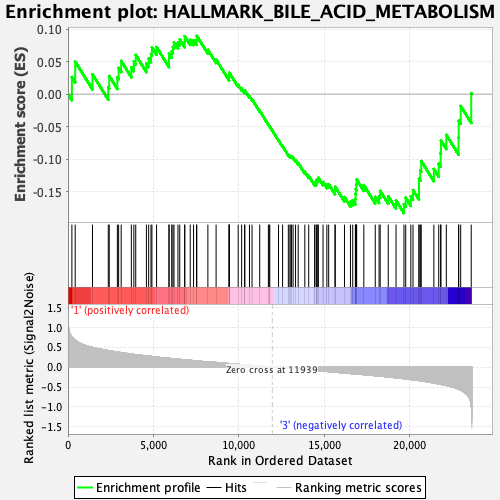
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group2_versus_Group4.GMP.mono_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.18324141 |
| Normalized Enrichment Score (NES) | -0.78717744 |
| Nominal p-value | 0.860735 |
| FDR q-value | 0.9973616 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Akr1d1 | 227 | 0.754 | 0.0262 | No |
| 2 | Gclm | 421 | 0.674 | 0.0501 | No |
| 3 | Bmp6 | 1433 | 0.492 | 0.0305 | No |
| 4 | Pex6 | 2356 | 0.410 | 0.0108 | No |
| 5 | Idi1 | 2413 | 0.408 | 0.0278 | No |
| 6 | Acsl5 | 2887 | 0.378 | 0.0257 | No |
| 7 | Pex16 | 2961 | 0.370 | 0.0402 | No |
| 8 | Retsat | 3111 | 0.361 | 0.0510 | No |
| 9 | Dhcr24 | 3706 | 0.326 | 0.0413 | No |
| 10 | Tfcp2l1 | 3853 | 0.316 | 0.0501 | No |
| 11 | Ephx2 | 3963 | 0.309 | 0.0602 | No |
| 12 | Gc | 4587 | 0.281 | 0.0471 | No |
| 13 | Dio2 | 4719 | 0.274 | 0.0546 | No |
| 14 | Scp2 | 4855 | 0.268 | 0.0616 | No |
| 15 | Lck | 4909 | 0.265 | 0.0719 | No |
| 16 | Npc1 | 5181 | 0.250 | 0.0723 | No |
| 17 | Abca8b | 5909 | 0.220 | 0.0519 | No |
| 18 | Fdxr | 5910 | 0.219 | 0.0623 | No |
| 19 | Klf1 | 6066 | 0.211 | 0.0658 | No |
| 20 | Pex11g | 6138 | 0.207 | 0.0726 | No |
| 21 | Nr3c2 | 6201 | 0.205 | 0.0797 | No |
| 22 | Gnpat | 6433 | 0.198 | 0.0793 | No |
| 23 | Idh1 | 6538 | 0.193 | 0.0841 | No |
| 24 | Aqp9 | 6828 | 0.182 | 0.0804 | No |
| 25 | Slc27a5 | 6830 | 0.182 | 0.0891 | No |
| 26 | Abcd3 | 7150 | 0.169 | 0.0835 | No |
| 27 | Isoc1 | 7345 | 0.161 | 0.0830 | No |
| 28 | Slc23a2 | 7518 | 0.153 | 0.0829 | No |
| 29 | Aldh9a1 | 7535 | 0.153 | 0.0895 | No |
| 30 | Cyp46a1 | 8183 | 0.127 | 0.0681 | No |
| 31 | Rbp1 | 8664 | 0.115 | 0.0532 | No |
| 32 | Hacl1 | 9423 | 0.088 | 0.0251 | No |
| 33 | Pxmp2 | 9425 | 0.088 | 0.0292 | No |
| 34 | Paox | 9438 | 0.087 | 0.0329 | No |
| 35 | Pex1 | 9956 | 0.068 | 0.0141 | No |
| 36 | Fads1 | 10158 | 0.061 | 0.0085 | No |
| 37 | Rxra | 10332 | 0.055 | 0.0037 | No |
| 38 | Soat2 | 10345 | 0.054 | 0.0058 | No |
| 39 | Pex11a | 10616 | 0.045 | -0.0035 | No |
| 40 | Pnpla8 | 10766 | 0.040 | -0.0080 | No |
| 41 | Nr1h4 | 11216 | 0.025 | -0.0259 | No |
| 42 | Ar | 11723 | 0.007 | -0.0470 | No |
| 43 | Lonp2 | 11767 | 0.006 | -0.0486 | No |
| 44 | Abca6 | 11805 | 0.005 | -0.0499 | No |
| 45 | Acsl1 | 12316 | -0.010 | -0.0711 | No |
| 46 | Hsd17b11 | 12554 | -0.018 | -0.0803 | No |
| 47 | Pex26 | 12890 | -0.030 | -0.0932 | No |
| 48 | Slc23a1 | 12981 | -0.033 | -0.0954 | No |
| 49 | Dio1 | 13042 | -0.035 | -0.0963 | No |
| 50 | Pex19 | 13081 | -0.036 | -0.0962 | No |
| 51 | Crot | 13169 | -0.039 | -0.0980 | No |
| 52 | Slc35b2 | 13312 | -0.045 | -0.1019 | No |
| 53 | Nudt12 | 13466 | -0.050 | -0.1061 | No |
| 54 | Pex7 | 13855 | -0.063 | -0.1196 | No |
| 55 | Cyp27a1 | 14082 | -0.070 | -0.1258 | No |
| 56 | Cyp39a1 | 14425 | -0.084 | -0.1364 | No |
| 57 | Idh2 | 14522 | -0.088 | -0.1363 | No |
| 58 | Abcd1 | 14524 | -0.088 | -0.1322 | No |
| 59 | Abcg4 | 14607 | -0.091 | -0.1314 | No |
| 60 | Hsd17b4 | 14647 | -0.092 | -0.1287 | No |
| 61 | Amacr | 14920 | -0.102 | -0.1354 | No |
| 62 | Sod1 | 15144 | -0.111 | -0.1396 | No |
| 63 | Gstk1 | 15250 | -0.115 | -0.1386 | No |
| 64 | Pfkm | 15619 | -0.128 | -0.1481 | No |
| 65 | Abca4 | 15635 | -0.129 | -0.1427 | No |
| 66 | Aldh1a1 | 16175 | -0.148 | -0.1585 | No |
| 67 | Mlycd | 16524 | -0.161 | -0.1656 | No |
| 68 | Efhc1 | 16658 | -0.167 | -0.1633 | No |
| 69 | Cat | 16812 | -0.173 | -0.1616 | No |
| 70 | Bcar3 | 16823 | -0.173 | -0.1538 | No |
| 71 | Abca5 | 16842 | -0.174 | -0.1463 | No |
| 72 | Pipox | 16874 | -0.176 | -0.1392 | No |
| 73 | Optn | 16888 | -0.176 | -0.1314 | No |
| 74 | Abca3 | 17306 | -0.190 | -0.1401 | No |
| 75 | Slc29a1 | 17975 | -0.216 | -0.1582 | No |
| 76 | Abcd2 | 18199 | -0.224 | -0.1570 | No |
| 77 | Fads2 | 18263 | -0.227 | -0.1489 | No |
| 78 | Abca2 | 18735 | -0.248 | -0.1571 | No |
| 79 | Slc22a18 | 19191 | -0.270 | -0.1636 | No |
| 80 | Lipe | 19654 | -0.291 | -0.1694 | Yes |
| 81 | Cyp7b1 | 19757 | -0.298 | -0.1596 | Yes |
| 82 | Abca9 | 20056 | -0.312 | -0.1574 | Yes |
| 83 | Abca1 | 20184 | -0.318 | -0.1476 | Yes |
| 84 | Hao1 | 20533 | -0.338 | -0.1463 | Yes |
| 85 | Pex13 | 20534 | -0.338 | -0.1303 | Yes |
| 86 | Sult2b1 | 20612 | -0.344 | -0.1172 | Yes |
| 87 | Gnmt | 20666 | -0.347 | -0.1029 | Yes |
| 88 | Nedd4 | 21404 | -0.399 | -0.1152 | Yes |
| 89 | Phyh | 21689 | -0.423 | -0.1072 | Yes |
| 90 | Slc27a2 | 21798 | -0.433 | -0.0912 | Yes |
| 91 | Prdx5 | 21818 | -0.434 | -0.0714 | Yes |
| 92 | Nr0b2 | 22133 | -0.460 | -0.0628 | Yes |
| 93 | Pecr | 22849 | -0.551 | -0.0670 | Yes |
| 94 | Atxn1 | 22858 | -0.553 | -0.0410 | Yes |
| 95 | Pex12 | 22973 | -0.579 | -0.0183 | Yes |
| 96 | Hsd3b7 | 23590 | -0.966 | 0.0014 | Yes |