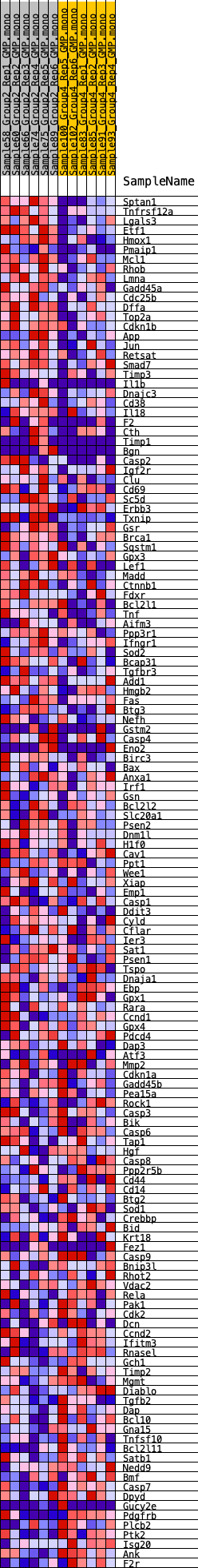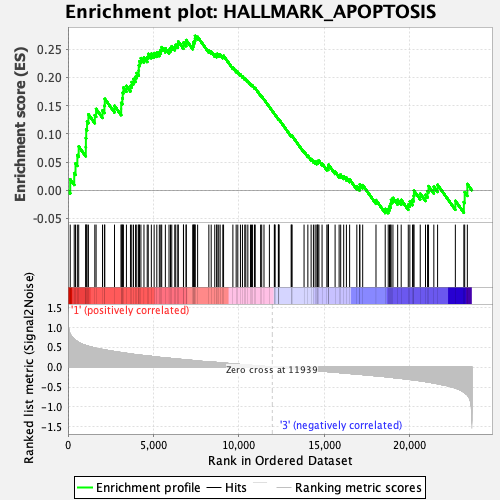
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group2_versus_Group4.GMP.mono_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_APOPTOSIS |
| Enrichment Score (ES) | 0.27355963 |
| Normalized Enrichment Score (NES) | 1.2248588 |
| Nominal p-value | 0.16475096 |
| FDR q-value | 0.5539922 |
| FWER p-Value | 0.863 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Sptan1 | 131 | 0.817 | 0.0193 | Yes |
| 2 | Tnfrsf12a | 364 | 0.694 | 0.0305 | Yes |
| 3 | Lgals3 | 440 | 0.669 | 0.0476 | Yes |
| 4 | Etf1 | 547 | 0.634 | 0.0624 | Yes |
| 5 | Hmox1 | 626 | 0.610 | 0.0776 | Yes |
| 6 | Pmaip1 | 1041 | 0.539 | 0.0764 | Yes |
| 7 | Mcl1 | 1047 | 0.538 | 0.0925 | Yes |
| 8 | Rhob | 1065 | 0.536 | 0.1081 | Yes |
| 9 | Lmna | 1115 | 0.527 | 0.1221 | Yes |
| 10 | Gadd45a | 1194 | 0.519 | 0.1345 | Yes |
| 11 | Cdc25b | 1569 | 0.475 | 0.1330 | Yes |
| 12 | Dffa | 1643 | 0.467 | 0.1441 | Yes |
| 13 | Top2a | 2019 | 0.440 | 0.1415 | Yes |
| 14 | Cdkn1b | 2130 | 0.430 | 0.1499 | Yes |
| 15 | App | 2153 | 0.428 | 0.1620 | Yes |
| 16 | Jun | 2721 | 0.387 | 0.1496 | Yes |
| 17 | Retsat | 3111 | 0.361 | 0.1441 | Yes |
| 18 | Smad7 | 3112 | 0.361 | 0.1550 | Yes |
| 19 | Timp3 | 3180 | 0.356 | 0.1630 | Yes |
| 20 | Il1b | 3201 | 0.356 | 0.1730 | Yes |
| 21 | Dnajc3 | 3241 | 0.353 | 0.1821 | Yes |
| 22 | Cd38 | 3418 | 0.342 | 0.1850 | Yes |
| 23 | Il18 | 3654 | 0.329 | 0.1850 | Yes |
| 24 | F2 | 3729 | 0.325 | 0.1917 | Yes |
| 25 | Cth | 3830 | 0.317 | 0.1971 | Yes |
| 26 | Timp1 | 3952 | 0.310 | 0.2013 | Yes |
| 27 | Bgn | 4022 | 0.308 | 0.2078 | Yes |
| 28 | Casp2 | 4137 | 0.303 | 0.2121 | Yes |
| 29 | Igf2r | 4141 | 0.302 | 0.2212 | Yes |
| 30 | Clu | 4182 | 0.300 | 0.2286 | Yes |
| 31 | Cd69 | 4265 | 0.295 | 0.2341 | Yes |
| 32 | Sc5d | 4444 | 0.289 | 0.2353 | Yes |
| 33 | Erbb3 | 4634 | 0.280 | 0.2358 | Yes |
| 34 | Txnip | 4694 | 0.276 | 0.2417 | Yes |
| 35 | Gsr | 4876 | 0.267 | 0.2421 | Yes |
| 36 | Brca1 | 5039 | 0.257 | 0.2430 | Yes |
| 37 | Sqstm1 | 5196 | 0.249 | 0.2439 | Yes |
| 38 | Gpx3 | 5343 | 0.242 | 0.2451 | Yes |
| 39 | Lef1 | 5428 | 0.237 | 0.2487 | Yes |
| 40 | Madd | 5482 | 0.235 | 0.2536 | Yes |
| 41 | Ctnnb1 | 5696 | 0.228 | 0.2515 | Yes |
| 42 | Fdxr | 5910 | 0.219 | 0.2491 | Yes |
| 43 | Bcl2l1 | 5996 | 0.215 | 0.2520 | Yes |
| 44 | Tnf | 6071 | 0.211 | 0.2553 | Yes |
| 45 | Aifm3 | 6247 | 0.203 | 0.2540 | Yes |
| 46 | Ppp3r1 | 6304 | 0.201 | 0.2577 | Yes |
| 47 | Ifngr1 | 6427 | 0.198 | 0.2586 | Yes |
| 48 | Sod2 | 6451 | 0.197 | 0.2636 | Yes |
| 49 | Bcap31 | 6754 | 0.185 | 0.2563 | Yes |
| 50 | Tgfbr3 | 6759 | 0.185 | 0.2618 | Yes |
| 51 | Add1 | 6913 | 0.179 | 0.2607 | Yes |
| 52 | Hmgb2 | 6914 | 0.179 | 0.2662 | Yes |
| 53 | Fas | 7301 | 0.163 | 0.2547 | Yes |
| 54 | Btg3 | 7326 | 0.162 | 0.2586 | Yes |
| 55 | Nefh | 7346 | 0.161 | 0.2627 | Yes |
| 56 | Gstm2 | 7408 | 0.158 | 0.2649 | Yes |
| 57 | Casp4 | 7428 | 0.158 | 0.2689 | Yes |
| 58 | Eno2 | 7432 | 0.158 | 0.2736 | Yes |
| 59 | Birc3 | 7583 | 0.151 | 0.2718 | No |
| 60 | Bax | 8240 | 0.125 | 0.2476 | No |
| 61 | Anxa1 | 8377 | 0.123 | 0.2456 | No |
| 62 | Irf1 | 8586 | 0.118 | 0.2403 | No |
| 63 | Gsn | 8697 | 0.114 | 0.2391 | No |
| 64 | Bcl2l2 | 8718 | 0.114 | 0.2417 | No |
| 65 | Slc20a1 | 8817 | 0.110 | 0.2409 | No |
| 66 | Psen2 | 8901 | 0.107 | 0.2406 | No |
| 67 | Dnm1l | 9058 | 0.101 | 0.2370 | No |
| 68 | H1f0 | 9101 | 0.099 | 0.2383 | No |
| 69 | Cav1 | 9646 | 0.079 | 0.2175 | No |
| 70 | Ppt1 | 9838 | 0.072 | 0.2116 | No |
| 71 | Wee1 | 9934 | 0.069 | 0.2096 | No |
| 72 | Xiap | 10089 | 0.063 | 0.2050 | No |
| 73 | Emp1 | 10206 | 0.059 | 0.2018 | No |
| 74 | Casp1 | 10342 | 0.054 | 0.1977 | No |
| 75 | Ddit3 | 10402 | 0.052 | 0.1968 | No |
| 76 | Cyld | 10521 | 0.048 | 0.1932 | No |
| 77 | Cflar | 10676 | 0.043 | 0.1880 | No |
| 78 | Ier3 | 10737 | 0.041 | 0.1867 | No |
| 79 | Sat1 | 10797 | 0.039 | 0.1853 | No |
| 80 | Psen1 | 10910 | 0.035 | 0.1816 | No |
| 81 | Tspo | 10952 | 0.034 | 0.1809 | No |
| 82 | Dnaja1 | 11269 | 0.023 | 0.1682 | No |
| 83 | Ebp | 11332 | 0.021 | 0.1661 | No |
| 84 | Gpx1 | 11466 | 0.016 | 0.1610 | No |
| 85 | Rara | 11782 | 0.005 | 0.1477 | No |
| 86 | Ccnd1 | 12071 | -0.001 | 0.1355 | No |
| 87 | Gpx4 | 12085 | -0.001 | 0.1350 | No |
| 88 | Pdcd4 | 12117 | -0.002 | 0.1337 | No |
| 89 | Dap3 | 12307 | -0.010 | 0.1260 | No |
| 90 | Atf3 | 12314 | -0.010 | 0.1260 | No |
| 91 | Mmp2 | 12338 | -0.011 | 0.1254 | No |
| 92 | Cdkn1a | 12342 | -0.011 | 0.1256 | No |
| 93 | Gadd45b | 13055 | -0.035 | 0.0963 | No |
| 94 | Pea15a | 13085 | -0.036 | 0.0962 | No |
| 95 | Rock1 | 13107 | -0.037 | 0.0964 | No |
| 96 | Casp3 | 13810 | -0.061 | 0.0684 | No |
| 97 | Bik | 14032 | -0.068 | 0.0611 | No |
| 98 | Casp6 | 14223 | -0.076 | 0.0553 | No |
| 99 | Tap1 | 14358 | -0.081 | 0.0520 | No |
| 100 | Hgf | 14458 | -0.085 | 0.0504 | No |
| 101 | Casp8 | 14559 | -0.089 | 0.0489 | No |
| 102 | Ppp2r5b | 14606 | -0.090 | 0.0497 | No |
| 103 | Cd44 | 14616 | -0.091 | 0.0520 | No |
| 104 | Cd14 | 14679 | -0.093 | 0.0522 | No |
| 105 | Btg2 | 14863 | -0.099 | 0.0474 | No |
| 106 | Sod1 | 15144 | -0.111 | 0.0389 | No |
| 107 | Crebbp | 15219 | -0.113 | 0.0392 | No |
| 108 | Bid | 15225 | -0.114 | 0.0424 | No |
| 109 | Krt18 | 15240 | -0.114 | 0.0453 | No |
| 110 | Fez1 | 15626 | -0.128 | 0.0328 | No |
| 111 | Casp9 | 15861 | -0.137 | 0.0270 | No |
| 112 | Bnip3l | 15953 | -0.141 | 0.0274 | No |
| 113 | Rhot2 | 16128 | -0.147 | 0.0244 | No |
| 114 | Vdac2 | 16288 | -0.152 | 0.0223 | No |
| 115 | Rela | 16476 | -0.159 | 0.0192 | No |
| 116 | Pak1 | 16897 | -0.177 | 0.0067 | No |
| 117 | Cdk2 | 17064 | -0.183 | 0.0052 | No |
| 118 | Dcn | 17072 | -0.184 | 0.0105 | No |
| 119 | Ccnd2 | 17240 | -0.187 | 0.0091 | No |
| 120 | Ifitm3 | 18018 | -0.217 | -0.0174 | No |
| 121 | Rnasel | 18558 | -0.239 | -0.0331 | No |
| 122 | Gch1 | 18734 | -0.248 | -0.0330 | No |
| 123 | Timp2 | 18808 | -0.251 | -0.0285 | No |
| 124 | Mgmt | 18875 | -0.253 | -0.0236 | No |
| 125 | Diablo | 18901 | -0.255 | -0.0169 | No |
| 126 | Tgfb2 | 19010 | -0.261 | -0.0136 | No |
| 127 | Dap | 19286 | -0.275 | -0.0169 | No |
| 128 | Bcl10 | 19493 | -0.283 | -0.0171 | No |
| 129 | Gna15 | 19907 | -0.307 | -0.0253 | No |
| 130 | Tnfsf10 | 19999 | -0.310 | -0.0198 | No |
| 131 | Bcl2l11 | 20156 | -0.316 | -0.0168 | No |
| 132 | Satb1 | 20227 | -0.320 | -0.0101 | No |
| 133 | Nedd9 | 20238 | -0.321 | -0.0007 | No |
| 134 | Bmf | 20607 | -0.343 | -0.0060 | No |
| 135 | Casp7 | 20915 | -0.363 | -0.0080 | No |
| 136 | Dpyd | 21035 | -0.372 | -0.0017 | No |
| 137 | Gucy2e | 21092 | -0.376 | 0.0073 | No |
| 138 | Pdgfrb | 21402 | -0.399 | 0.0063 | No |
| 139 | Plcb2 | 21622 | -0.417 | 0.0096 | No |
| 140 | Ptk2 | 22661 | -0.521 | -0.0187 | No |
| 141 | Isg20 | 23159 | -0.624 | -0.0209 | No |
| 142 | Ank | 23208 | -0.641 | -0.0034 | No |
| 143 | F2r | 23363 | -0.694 | 0.0111 | No |