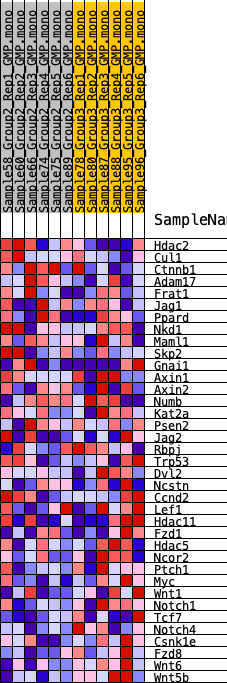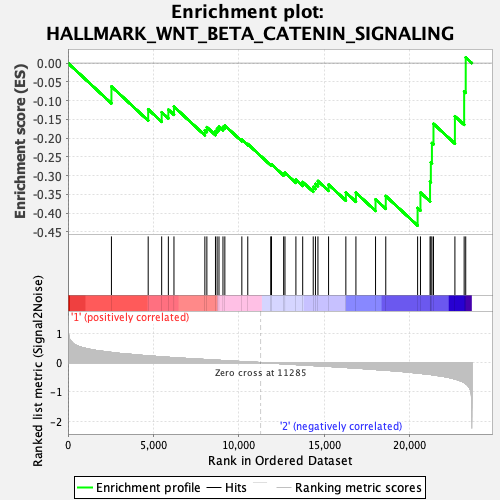
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group2_versus_Group3.GMP.mono_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | GMP.mono_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_WNT_BETA_CATENIN_SIGNALING |
| Enrichment Score (ES) | -0.43351898 |
| Normalized Enrichment Score (NES) | -1.2667422 |
| Nominal p-value | 0.23640168 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.843 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hdac2 | 2541 | 0.344 | -0.0624 | No |
| 2 | Cul1 | 4691 | 0.229 | -0.1233 | No |
| 3 | Ctnnb1 | 5480 | 0.194 | -0.1312 | No |
| 4 | Adam17 | 5871 | 0.178 | -0.1242 | No |
| 5 | Frat1 | 6199 | 0.166 | -0.1162 | No |
| 6 | Jag1 | 8007 | 0.103 | -0.1792 | No |
| 7 | Ppard | 8124 | 0.099 | -0.1710 | No |
| 8 | Nkd1 | 8627 | 0.082 | -0.1814 | No |
| 9 | Maml1 | 8724 | 0.079 | -0.1750 | No |
| 10 | Skp2 | 8833 | 0.076 | -0.1696 | No |
| 11 | Gnai1 | 9066 | 0.068 | -0.1705 | No |
| 12 | Axin1 | 9176 | 0.065 | -0.1666 | No |
| 13 | Axin2 | 10170 | 0.034 | -0.2042 | No |
| 14 | Numb | 10517 | 0.023 | -0.2158 | No |
| 15 | Kat2a | 11860 | -0.017 | -0.2705 | No |
| 16 | Psen2 | 11906 | -0.018 | -0.2700 | No |
| 17 | Jag2 | 12616 | -0.040 | -0.2948 | No |
| 18 | Rbpj | 12693 | -0.042 | -0.2924 | No |
| 19 | Trp53 | 13333 | -0.065 | -0.3109 | No |
| 20 | Dvl2 | 13730 | -0.079 | -0.3173 | No |
| 21 | Ncstn | 14346 | -0.099 | -0.3304 | No |
| 22 | Ccnd2 | 14477 | -0.102 | -0.3224 | No |
| 23 | Lef1 | 14622 | -0.106 | -0.3146 | No |
| 24 | Hdac11 | 15244 | -0.128 | -0.3241 | No |
| 25 | Fzd1 | 16255 | -0.165 | -0.3451 | No |
| 26 | Hdac5 | 16844 | -0.185 | -0.3457 | No |
| 27 | Ncor2 | 17992 | -0.234 | -0.3635 | No |
| 28 | Ptch1 | 18590 | -0.259 | -0.3546 | No |
| 29 | Myc | 20453 | -0.356 | -0.3866 | Yes |
| 30 | Wnt1 | 20620 | -0.365 | -0.3455 | Yes |
| 31 | Notch1 | 21181 | -0.402 | -0.3162 | Yes |
| 32 | Tcf7 | 21228 | -0.405 | -0.2648 | Yes |
| 33 | Notch4 | 21291 | -0.410 | -0.2133 | Yes |
| 34 | Csnk1e | 21382 | -0.419 | -0.1619 | Yes |
| 35 | Fzd8 | 22635 | -0.555 | -0.1419 | Yes |
| 36 | Wnt6 | 23176 | -0.676 | -0.0756 | Yes |
| 37 | Wnt5b | 23268 | -0.717 | 0.0151 | Yes |