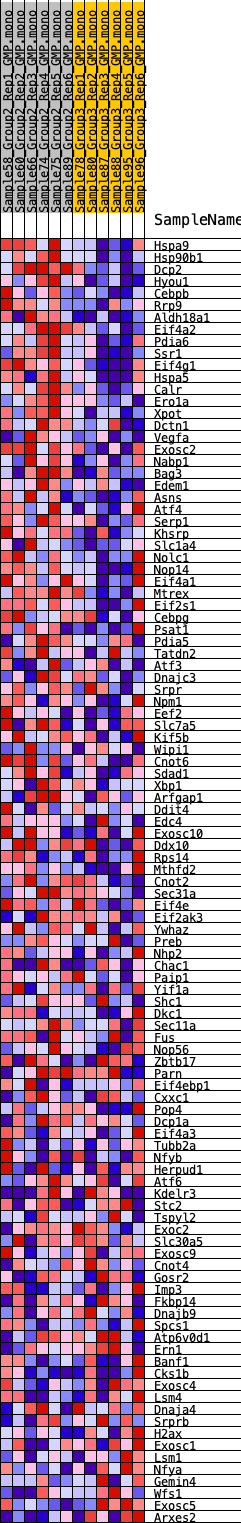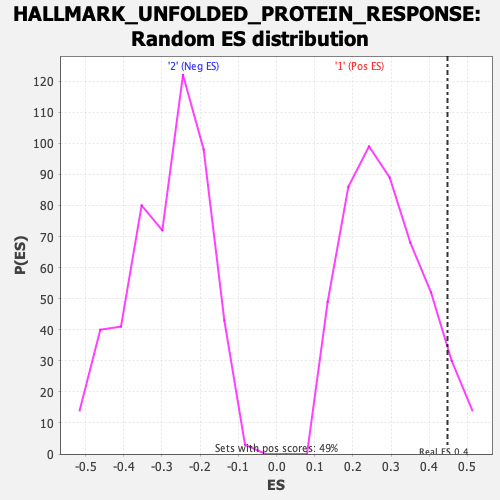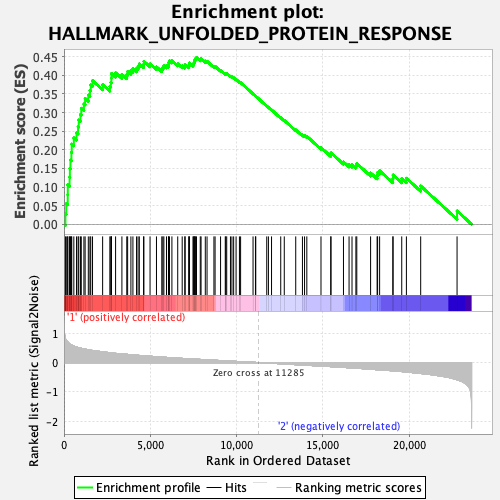
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group2_versus_Group3.GMP.mono_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | GMP.mono_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.44783357 |
| Normalized Enrichment Score (NES) | 1.5641392 |
| Nominal p-value | 0.061601643 |
| FDR q-value | 0.1792314 |
| FWER p-Value | 0.323 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hspa9 | 83 | 0.816 | 0.0281 | Yes |
| 2 | Hsp90b1 | 131 | 0.761 | 0.0556 | Yes |
| 3 | Dcp2 | 215 | 0.699 | 0.0792 | Yes |
| 4 | Hyou1 | 219 | 0.697 | 0.1061 | Yes |
| 5 | Cebpb | 324 | 0.636 | 0.1263 | Yes |
| 6 | Rrp9 | 340 | 0.628 | 0.1501 | Yes |
| 7 | Aldh18a1 | 374 | 0.613 | 0.1724 | Yes |
| 8 | Eif4a2 | 430 | 0.596 | 0.1932 | Yes |
| 9 | Pdia6 | 449 | 0.591 | 0.2153 | Yes |
| 10 | Ssr1 | 570 | 0.562 | 0.2320 | Yes |
| 11 | Eif4g1 | 726 | 0.525 | 0.2458 | Yes |
| 12 | Hspa5 | 816 | 0.513 | 0.2619 | Yes |
| 13 | Calr | 854 | 0.507 | 0.2799 | Yes |
| 14 | Ero1a | 953 | 0.491 | 0.2948 | Yes |
| 15 | Xpot | 1001 | 0.485 | 0.3116 | Yes |
| 16 | Dctn1 | 1154 | 0.463 | 0.3231 | Yes |
| 17 | Vegfa | 1224 | 0.456 | 0.3378 | Yes |
| 18 | Exosc2 | 1409 | 0.437 | 0.3470 | Yes |
| 19 | Nabp1 | 1503 | 0.427 | 0.3596 | Yes |
| 20 | Bag3 | 1550 | 0.423 | 0.3740 | Yes |
| 21 | Edem1 | 1653 | 0.411 | 0.3856 | Yes |
| 22 | Asns | 2239 | 0.363 | 0.3748 | Yes |
| 23 | Atf4 | 2658 | 0.336 | 0.3701 | Yes |
| 24 | Serp1 | 2712 | 0.332 | 0.3807 | Yes |
| 25 | Khsrp | 2745 | 0.329 | 0.3921 | Yes |
| 26 | Slc1a4 | 2749 | 0.329 | 0.4047 | Yes |
| 27 | Nolc1 | 2990 | 0.312 | 0.4066 | Yes |
| 28 | Nop14 | 3356 | 0.293 | 0.4025 | Yes |
| 29 | Eif4a1 | 3638 | 0.281 | 0.4014 | Yes |
| 30 | Mtrex | 3688 | 0.278 | 0.4101 | Yes |
| 31 | Eif2s1 | 3877 | 0.268 | 0.4125 | Yes |
| 32 | Cebpg | 3994 | 0.262 | 0.4177 | Yes |
| 33 | Psat1 | 4204 | 0.251 | 0.4186 | Yes |
| 34 | Pdia5 | 4291 | 0.246 | 0.4245 | Yes |
| 35 | Tatdn2 | 4365 | 0.242 | 0.4308 | Yes |
| 36 | Atf3 | 4612 | 0.230 | 0.4292 | Yes |
| 37 | Dnajc3 | 4629 | 0.230 | 0.4374 | Yes |
| 38 | Srpr | 4987 | 0.215 | 0.4306 | Yes |
| 39 | Npm1 | 5359 | 0.200 | 0.4226 | Yes |
| 40 | Eef2 | 5668 | 0.187 | 0.4167 | Yes |
| 41 | Slc7a5 | 5728 | 0.184 | 0.4213 | Yes |
| 42 | Kif5b | 5788 | 0.182 | 0.4259 | Yes |
| 43 | Wipi1 | 5940 | 0.176 | 0.4263 | Yes |
| 44 | Cnot6 | 6054 | 0.171 | 0.4281 | Yes |
| 45 | Sdad1 | 6065 | 0.171 | 0.4343 | Yes |
| 46 | Xbp1 | 6113 | 0.169 | 0.4389 | Yes |
| 47 | Arfgap1 | 6254 | 0.164 | 0.4393 | Yes |
| 48 | Ddit4 | 6591 | 0.151 | 0.4308 | Yes |
| 49 | Edc4 | 6850 | 0.142 | 0.4254 | Yes |
| 50 | Exosc10 | 7002 | 0.136 | 0.4242 | Yes |
| 51 | Ddx10 | 7020 | 0.135 | 0.4287 | Yes |
| 52 | Rps14 | 7231 | 0.127 | 0.4247 | Yes |
| 53 | Mthfd2 | 7252 | 0.126 | 0.4288 | Yes |
| 54 | Cnot2 | 7266 | 0.125 | 0.4331 | Yes |
| 55 | Sec31a | 7476 | 0.122 | 0.4289 | Yes |
| 56 | Eif4e | 7514 | 0.120 | 0.4320 | Yes |
| 57 | Eif2ak3 | 7542 | 0.119 | 0.4354 | Yes |
| 58 | Ywhaz | 7555 | 0.118 | 0.4395 | Yes |
| 59 | Preb | 7595 | 0.117 | 0.4424 | Yes |
| 60 | Nhp2 | 7629 | 0.116 | 0.4455 | Yes |
| 61 | Chac1 | 7678 | 0.114 | 0.4478 | Yes |
| 62 | Paip1 | 7897 | 0.107 | 0.4427 | No |
| 63 | Yif1a | 7943 | 0.105 | 0.4449 | No |
| 64 | Shc1 | 8189 | 0.097 | 0.4382 | No |
| 65 | Dkc1 | 8290 | 0.094 | 0.4376 | No |
| 66 | Sec11a | 8689 | 0.080 | 0.4238 | No |
| 67 | Fus | 8756 | 0.078 | 0.4240 | No |
| 68 | Nop56 | 9074 | 0.068 | 0.4132 | No |
| 69 | Zbtb17 | 9346 | 0.059 | 0.4039 | No |
| 70 | Parn | 9375 | 0.058 | 0.4050 | No |
| 71 | Eif4ebp1 | 9430 | 0.056 | 0.4049 | No |
| 72 | Cxxc1 | 9647 | 0.049 | 0.3976 | No |
| 73 | Pop4 | 9733 | 0.046 | 0.3958 | No |
| 74 | Dcp1a | 9832 | 0.044 | 0.3933 | No |
| 75 | Eif4a3 | 9973 | 0.039 | 0.3889 | No |
| 76 | Tubb2a | 10168 | 0.034 | 0.3819 | No |
| 77 | Nfyb | 10235 | 0.032 | 0.3803 | No |
| 78 | Herpud1 | 10956 | 0.010 | 0.3501 | No |
| 79 | Atf6 | 11096 | 0.005 | 0.3444 | No |
| 80 | Kdelr3 | 11114 | 0.005 | 0.3439 | No |
| 81 | Stc2 | 11750 | -0.014 | 0.3174 | No |
| 82 | Tspyl2 | 11847 | -0.017 | 0.3140 | No |
| 83 | Exoc2 | 12028 | -0.022 | 0.3071 | No |
| 84 | Slc30a5 | 12561 | -0.038 | 0.2860 | No |
| 85 | Exosc9 | 12766 | -0.045 | 0.2790 | No |
| 86 | Cnot4 | 13430 | -0.069 | 0.2535 | No |
| 87 | Gosr2 | 13819 | -0.082 | 0.2402 | No |
| 88 | Imp3 | 13941 | -0.086 | 0.2384 | No |
| 89 | Fkbp14 | 14073 | -0.090 | 0.2363 | No |
| 90 | Dnajb9 | 14894 | -0.116 | 0.2059 | No |
| 91 | Spcs1 | 15459 | -0.135 | 0.1872 | No |
| 92 | Atp6v0d1 | 15475 | -0.136 | 0.1918 | No |
| 93 | Ern1 | 16197 | -0.163 | 0.1674 | No |
| 94 | Banf1 | 16521 | -0.173 | 0.1604 | No |
| 95 | Cks1b | 16692 | -0.180 | 0.1602 | No |
| 96 | Exosc4 | 16917 | -0.188 | 0.1579 | No |
| 97 | Lsm4 | 16969 | -0.190 | 0.1631 | No |
| 98 | Dnaja4 | 17769 | -0.223 | 0.1378 | No |
| 99 | Srprb | 18147 | -0.240 | 0.1311 | No |
| 100 | H2ax | 18175 | -0.241 | 0.1393 | No |
| 101 | Exosc1 | 18292 | -0.246 | 0.1439 | No |
| 102 | Lsm1 | 19059 | -0.278 | 0.1221 | No |
| 103 | Nfya | 19071 | -0.279 | 0.1324 | No |
| 104 | Gemin4 | 19574 | -0.303 | 0.1228 | No |
| 105 | Wfs1 | 19844 | -0.318 | 0.1238 | No |
| 106 | Exosc5 | 20672 | -0.369 | 0.1029 | No |
| 107 | Arxes2 | 22780 | -0.583 | 0.0359 | No |