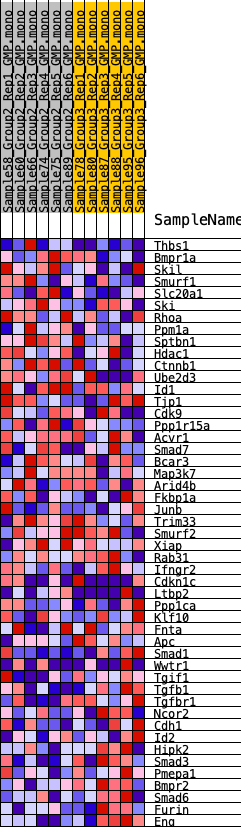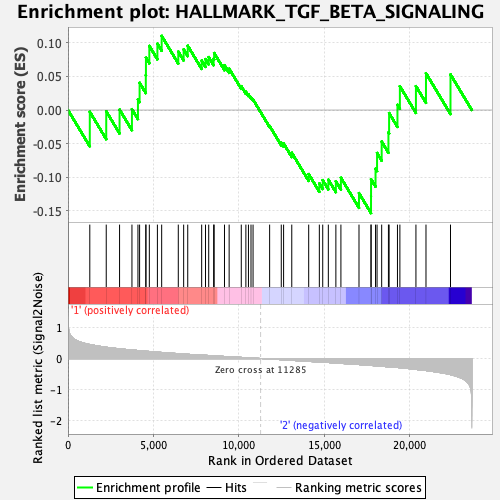
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group2_versus_Group3.GMP.mono_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | GMP.mono_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_TGF_BETA_SIGNALING |
| Enrichment Score (ES) | -0.15332034 |
| Normalized Enrichment Score (NES) | -0.6712045 |
| Nominal p-value | 0.8995984 |
| FDR q-value | 0.9591617 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Thbs1 | 1275 | 0.451 | -0.0026 | No |
| 2 | Bmpr1a | 2237 | 0.364 | -0.0019 | No |
| 3 | Skil | 3017 | 0.311 | 0.0006 | No |
| 4 | Smurf1 | 3741 | 0.275 | 0.0013 | No |
| 5 | Slc20a1 | 4092 | 0.257 | 0.0158 | No |
| 6 | Ski | 4189 | 0.252 | 0.0405 | No |
| 7 | Rhoa | 4552 | 0.232 | 0.0516 | No |
| 8 | Ppm1a | 4560 | 0.232 | 0.0778 | No |
| 9 | Sptbn1 | 4758 | 0.227 | 0.0954 | No |
| 10 | Hdac1 | 5230 | 0.204 | 0.0987 | No |
| 11 | Ctnnb1 | 5480 | 0.194 | 0.1103 | No |
| 12 | Ube2d3 | 6452 | 0.157 | 0.0869 | No |
| 13 | Id1 | 6766 | 0.144 | 0.0901 | No |
| 14 | Tjp1 | 7005 | 0.136 | 0.0956 | No |
| 15 | Cdk9 | 7820 | 0.110 | 0.0736 | No |
| 16 | Ppp1r15a | 8045 | 0.102 | 0.0757 | No |
| 17 | Acvr1 | 8232 | 0.095 | 0.0787 | No |
| 18 | Smad7 | 8520 | 0.086 | 0.0763 | No |
| 19 | Bcar3 | 8551 | 0.085 | 0.0848 | No |
| 20 | Map3k7 | 9155 | 0.065 | 0.0666 | No |
| 21 | Arid4b | 9427 | 0.056 | 0.0615 | No |
| 22 | Fkbp1a | 10137 | 0.035 | 0.0354 | No |
| 23 | Junb | 10406 | 0.027 | 0.0271 | No |
| 24 | Trim33 | 10557 | 0.022 | 0.0233 | No |
| 25 | Smurf2 | 10706 | 0.018 | 0.0191 | No |
| 26 | Xiap | 10826 | 0.014 | 0.0156 | No |
| 27 | Rab31 | 11796 | -0.015 | -0.0238 | No |
| 28 | Ifngr2 | 12477 | -0.036 | -0.0485 | No |
| 29 | Cdkn1c | 12613 | -0.039 | -0.0497 | No |
| 30 | Ltbp2 | 13091 | -0.056 | -0.0635 | No |
| 31 | Ppp1ca | 14081 | -0.090 | -0.0952 | No |
| 32 | Klf10 | 14704 | -0.109 | -0.1092 | No |
| 33 | Fnta | 14902 | -0.116 | -0.1042 | No |
| 34 | Apc | 15230 | -0.127 | -0.1035 | No |
| 35 | Smad1 | 15671 | -0.143 | -0.1059 | No |
| 36 | Wwtr1 | 15966 | -0.155 | -0.1007 | No |
| 37 | Tgif1 | 17024 | -0.193 | -0.1235 | No |
| 38 | Tgfb1 | 17728 | -0.221 | -0.1281 | Yes |
| 39 | Tgfbr1 | 17730 | -0.221 | -0.1029 | Yes |
| 40 | Ncor2 | 17992 | -0.234 | -0.0872 | Yes |
| 41 | Cdh1 | 18081 | -0.238 | -0.0638 | Yes |
| 42 | Id2 | 18351 | -0.249 | -0.0468 | Yes |
| 43 | Hipk2 | 18746 | -0.265 | -0.0333 | Yes |
| 44 | Smad3 | 18786 | -0.267 | -0.0045 | Yes |
| 45 | Pmepa1 | 19274 | -0.288 | 0.0077 | Yes |
| 46 | Bmpr2 | 19416 | -0.295 | 0.0354 | Yes |
| 47 | Smad6 | 20355 | -0.350 | 0.0355 | Yes |
| 48 | Furin | 20943 | -0.386 | 0.0547 | Yes |
| 49 | Eng | 22377 | -0.516 | 0.0529 | Yes |