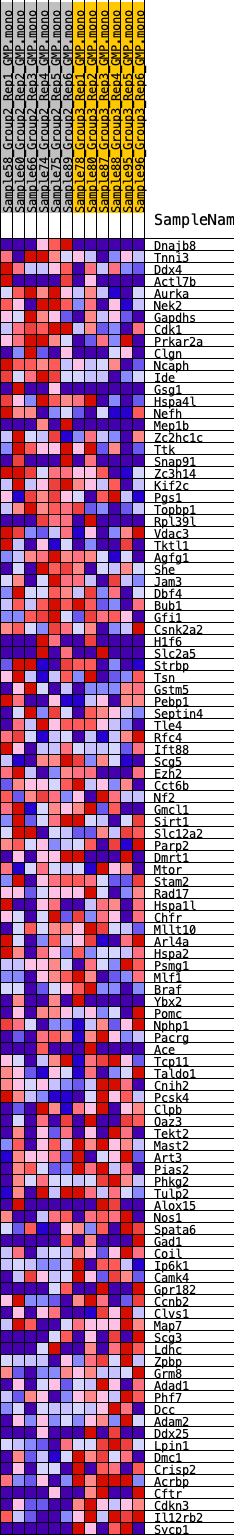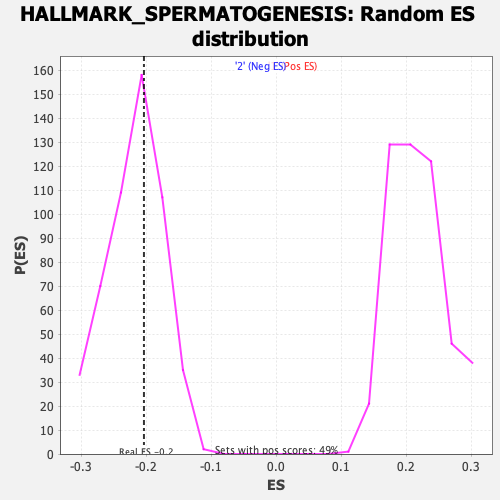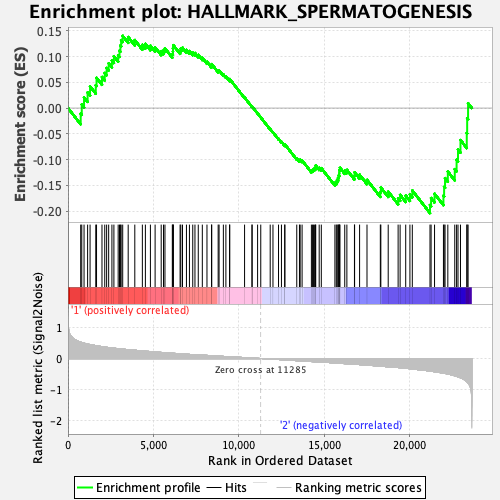
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group2_versus_Group3.GMP.mono_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | GMP.mono_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | -0.20389037 |
| Normalized Enrichment Score (NES) | -0.93479997 |
| Nominal p-value | 0.58949417 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.996 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dnajb8 | 743 | 0.523 | -0.0109 | No |
| 2 | Tnni3 | 805 | 0.515 | 0.0069 | No |
| 3 | Ddx4 | 939 | 0.492 | 0.0208 | No |
| 4 | Actl7b | 1147 | 0.464 | 0.0303 | No |
| 5 | Aurka | 1291 | 0.448 | 0.0420 | No |
| 6 | Nek2 | 1625 | 0.413 | 0.0442 | No |
| 7 | Gapdhs | 1669 | 0.409 | 0.0585 | No |
| 8 | Cdk1 | 1988 | 0.384 | 0.0602 | No |
| 9 | Prkar2a | 2150 | 0.370 | 0.0680 | No |
| 10 | Clgn | 2260 | 0.361 | 0.0777 | No |
| 11 | Ncaph | 2379 | 0.353 | 0.0867 | No |
| 12 | Ide | 2569 | 0.342 | 0.0922 | No |
| 13 | Gsg1 | 2685 | 0.334 | 0.1005 | No |
| 14 | Hspa4l | 2935 | 0.315 | 0.1024 | No |
| 15 | Nefh | 3013 | 0.311 | 0.1115 | No |
| 16 | Mep1b | 3066 | 0.308 | 0.1215 | No |
| 17 | Zc2hc1c | 3109 | 0.307 | 0.1318 | No |
| 18 | Ttk | 3200 | 0.303 | 0.1400 | No |
| 19 | Snap91 | 3521 | 0.287 | 0.1378 | No |
| 20 | Zc3h14 | 3908 | 0.267 | 0.1319 | No |
| 21 | Kif2c | 4350 | 0.243 | 0.1228 | No |
| 22 | Pgs1 | 4527 | 0.234 | 0.1245 | No |
| 23 | Topbp1 | 4821 | 0.224 | 0.1210 | No |
| 24 | Rpl39l | 5096 | 0.211 | 0.1176 | No |
| 25 | Vdac3 | 5452 | 0.196 | 0.1103 | No |
| 26 | Tktl1 | 5588 | 0.189 | 0.1121 | No |
| 27 | Agfg1 | 5672 | 0.187 | 0.1159 | No |
| 28 | She | 6103 | 0.169 | 0.1043 | No |
| 29 | Jam3 | 6137 | 0.168 | 0.1096 | No |
| 30 | Dbf4 | 6143 | 0.168 | 0.1160 | No |
| 31 | Bub1 | 6162 | 0.167 | 0.1219 | No |
| 32 | Gfi1 | 6558 | 0.153 | 0.1111 | No |
| 33 | Csnk2a2 | 6600 | 0.151 | 0.1154 | No |
| 34 | H1f6 | 6698 | 0.147 | 0.1171 | No |
| 35 | Slc2a5 | 6929 | 0.139 | 0.1128 | No |
| 36 | Strbp | 7104 | 0.132 | 0.1106 | No |
| 37 | Tsn | 7298 | 0.124 | 0.1073 | No |
| 38 | Gstm5 | 7427 | 0.123 | 0.1067 | No |
| 39 | Pebp1 | 7621 | 0.116 | 0.1031 | No |
| 40 | Septin4 | 7858 | 0.108 | 0.0974 | No |
| 41 | Tle4 | 8130 | 0.099 | 0.0898 | No |
| 42 | Rfc4 | 8398 | 0.090 | 0.0820 | No |
| 43 | Ift88 | 8408 | 0.090 | 0.0852 | No |
| 44 | Scg5 | 8785 | 0.077 | 0.0722 | No |
| 45 | Ezh2 | 8838 | 0.075 | 0.0730 | No |
| 46 | Cct6b | 9089 | 0.067 | 0.0651 | No |
| 47 | Nf2 | 9237 | 0.062 | 0.0613 | No |
| 48 | Gmcl1 | 9455 | 0.055 | 0.0542 | No |
| 49 | Sirt1 | 9460 | 0.055 | 0.0562 | No |
| 50 | Slc12a2 | 10330 | 0.029 | 0.0204 | No |
| 51 | Parp2 | 10765 | 0.016 | 0.0026 | No |
| 52 | Dmrt1 | 10769 | 0.016 | 0.0031 | No |
| 53 | Mtor | 11092 | 0.005 | -0.0104 | No |
| 54 | Stam2 | 11277 | 0.000 | -0.0182 | No |
| 55 | Rad17 | 11829 | -0.016 | -0.0410 | No |
| 56 | Hspa1l | 11991 | -0.021 | -0.0470 | No |
| 57 | Chfr | 12316 | -0.030 | -0.0596 | No |
| 58 | Mllt10 | 12487 | -0.036 | -0.0654 | No |
| 59 | Arl4a | 12681 | -0.042 | -0.0720 | No |
| 60 | Hspa2 | 12685 | -0.042 | -0.0704 | No |
| 61 | Psmg1 | 13383 | -0.067 | -0.0974 | No |
| 62 | Mlf1 | 13541 | -0.072 | -0.1012 | No |
| 63 | Braf | 13582 | -0.074 | -0.1000 | No |
| 64 | Ybx2 | 13695 | -0.078 | -0.1017 | No |
| 65 | Pomc | 14239 | -0.095 | -0.1210 | No |
| 66 | Nphp1 | 14308 | -0.098 | -0.1200 | No |
| 67 | Pacrg | 14359 | -0.099 | -0.1182 | No |
| 68 | Ace | 14427 | -0.101 | -0.1171 | No |
| 69 | Tcp11 | 14484 | -0.102 | -0.1154 | No |
| 70 | Taldo1 | 14489 | -0.102 | -0.1115 | No |
| 71 | Cnih2 | 14695 | -0.109 | -0.1159 | No |
| 72 | Pcsk4 | 14819 | -0.114 | -0.1167 | No |
| 73 | Clpb | 15615 | -0.141 | -0.1449 | No |
| 74 | Oaz3 | 15696 | -0.144 | -0.1426 | No |
| 75 | Tekt2 | 15748 | -0.146 | -0.1390 | No |
| 76 | Mast2 | 15803 | -0.149 | -0.1354 | No |
| 77 | Art3 | 15834 | -0.150 | -0.1307 | No |
| 78 | Pias2 | 15872 | -0.151 | -0.1263 | No |
| 79 | Phkg2 | 15874 | -0.151 | -0.1204 | No |
| 80 | Tulp2 | 15911 | -0.153 | -0.1159 | No |
| 81 | Alox15 | 16190 | -0.162 | -0.1213 | No |
| 82 | Nos1 | 16315 | -0.167 | -0.1199 | No |
| 83 | Spata6 | 16758 | -0.181 | -0.1315 | No |
| 84 | Gad1 | 16767 | -0.182 | -0.1247 | No |
| 85 | Coil | 17059 | -0.194 | -0.1294 | No |
| 86 | Ip6k1 | 17492 | -0.212 | -0.1393 | No |
| 87 | Camk4 | 18272 | -0.245 | -0.1628 | No |
| 88 | Gpr182 | 18302 | -0.246 | -0.1542 | No |
| 89 | Ccnb2 | 18733 | -0.264 | -0.1621 | No |
| 90 | Clvs1 | 19313 | -0.290 | -0.1752 | No |
| 91 | Map7 | 19430 | -0.295 | -0.1685 | No |
| 92 | Scg3 | 19757 | -0.314 | -0.1699 | No |
| 93 | Ldhc | 20003 | -0.327 | -0.1673 | No |
| 94 | Zpbp | 20143 | -0.337 | -0.1599 | No |
| 95 | Grm8 | 21178 | -0.402 | -0.1880 | Yes |
| 96 | Adad1 | 21242 | -0.406 | -0.1746 | Yes |
| 97 | Phf7 | 21444 | -0.425 | -0.1663 | Yes |
| 98 | Dcc | 21964 | -0.469 | -0.1698 | Yes |
| 99 | Adam2 | 22004 | -0.474 | -0.1527 | Yes |
| 100 | Ddx25 | 22058 | -0.479 | -0.1360 | Yes |
| 101 | Lpin1 | 22221 | -0.496 | -0.1233 | Yes |
| 102 | Dmc1 | 22627 | -0.554 | -0.1186 | Yes |
| 103 | Crisp2 | 22734 | -0.572 | -0.1004 | Yes |
| 104 | Acrbp | 22816 | -0.590 | -0.0805 | Yes |
| 105 | Cftr | 22961 | -0.618 | -0.0622 | Yes |
| 106 | Cdkn3 | 23329 | -0.742 | -0.0484 | Yes |
| 107 | Il12rb2 | 23353 | -0.752 | -0.0196 | Yes |
| 108 | Sycp1 | 23406 | -0.785 | 0.0093 | Yes |