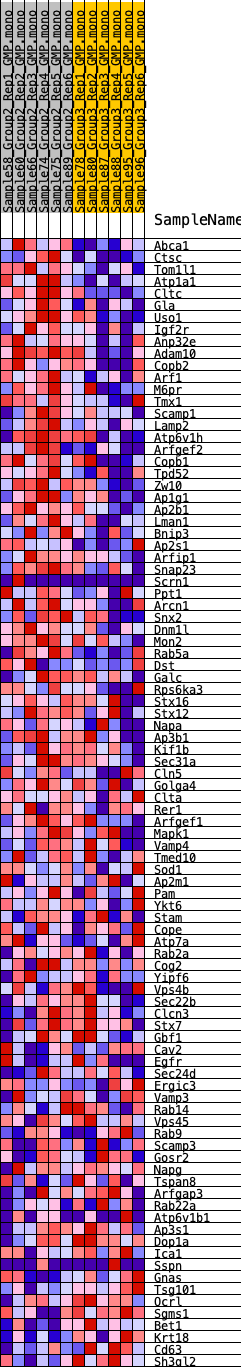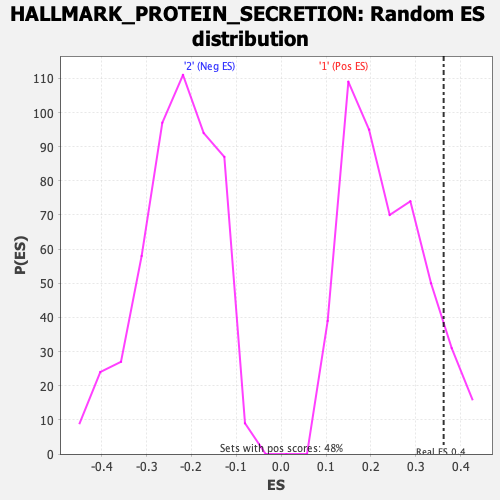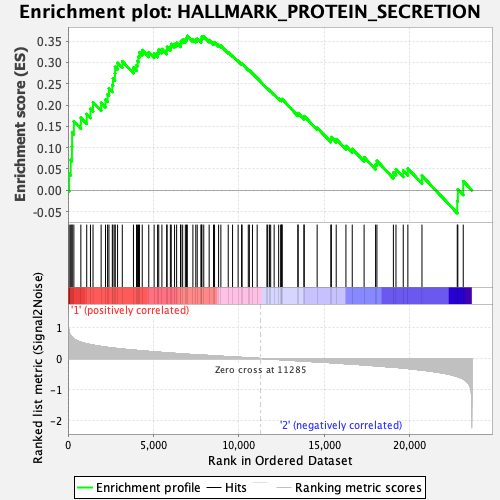
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group2_versus_Group3.GMP.mono_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | GMP.mono_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | 0.36234146 |
| Normalized Enrichment Score (NES) | 1.5651948 |
| Nominal p-value | 0.08884297 |
| FDR q-value | 0.22330754 |
| FWER p-Value | 0.323 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Abca1 | 68 | 0.844 | 0.0387 | Yes |
| 2 | Ctsc | 158 | 0.736 | 0.0711 | Yes |
| 3 | Tom1l1 | 229 | 0.693 | 0.1023 | Yes |
| 4 | Atp1a1 | 241 | 0.683 | 0.1354 | Yes |
| 5 | Cltc | 343 | 0.626 | 0.1620 | Yes |
| 6 | Gla | 752 | 0.523 | 0.1704 | Yes |
| 7 | Uso1 | 1095 | 0.470 | 0.1789 | Yes |
| 8 | Igf2r | 1314 | 0.445 | 0.1916 | Yes |
| 9 | Anp32e | 1464 | 0.432 | 0.2065 | Yes |
| 10 | Adam10 | 1938 | 0.386 | 0.2054 | Yes |
| 11 | Copb2 | 2195 | 0.367 | 0.2126 | Yes |
| 12 | Arf1 | 2316 | 0.356 | 0.2250 | Yes |
| 13 | M6pr | 2395 | 0.352 | 0.2391 | Yes |
| 14 | Tmx1 | 2592 | 0.340 | 0.2475 | Yes |
| 15 | Scamp1 | 2632 | 0.338 | 0.2625 | Yes |
| 16 | Lamp2 | 2746 | 0.329 | 0.2739 | Yes |
| 17 | Atp6v1h | 2756 | 0.328 | 0.2896 | Yes |
| 18 | Arfgef2 | 2900 | 0.318 | 0.2992 | Yes |
| 19 | Copb1 | 3176 | 0.304 | 0.3025 | Yes |
| 20 | Tpd52 | 3829 | 0.270 | 0.2881 | Yes |
| 21 | Zw10 | 4009 | 0.261 | 0.2933 | Yes |
| 22 | Ap1g1 | 4070 | 0.258 | 0.3035 | Yes |
| 23 | Ap2b1 | 4125 | 0.255 | 0.3137 | Yes |
| 24 | Lman1 | 4181 | 0.252 | 0.3238 | Yes |
| 25 | Bnip3 | 4343 | 0.244 | 0.3290 | Yes |
| 26 | Ap2s1 | 4725 | 0.228 | 0.3240 | Yes |
| 27 | Arfip1 | 5039 | 0.213 | 0.3212 | Yes |
| 28 | Snap23 | 5242 | 0.204 | 0.3226 | Yes |
| 29 | Scrn1 | 5309 | 0.201 | 0.3297 | Yes |
| 30 | Ppt1 | 5494 | 0.193 | 0.3314 | Yes |
| 31 | Arcn1 | 5776 | 0.182 | 0.3285 | Yes |
| 32 | Snx2 | 5795 | 0.181 | 0.3366 | Yes |
| 33 | Dnm1l | 5989 | 0.174 | 0.3370 | Yes |
| 34 | Mon2 | 6039 | 0.172 | 0.3434 | Yes |
| 35 | Rab5a | 6234 | 0.165 | 0.3432 | Yes |
| 36 | Dst | 6355 | 0.160 | 0.3460 | Yes |
| 37 | Galc | 6581 | 0.152 | 0.3439 | Yes |
| 38 | Rps6ka3 | 6608 | 0.151 | 0.3502 | Yes |
| 39 | Stx16 | 6708 | 0.147 | 0.3533 | Yes |
| 40 | Stx12 | 6865 | 0.141 | 0.3536 | Yes |
| 41 | Napa | 6931 | 0.139 | 0.3576 | Yes |
| 42 | Ap3b1 | 6980 | 0.137 | 0.3623 | Yes |
| 43 | Kif1b | 7307 | 0.124 | 0.3546 | No |
| 44 | Sec31a | 7476 | 0.122 | 0.3534 | No |
| 45 | Cln5 | 7560 | 0.118 | 0.3557 | No |
| 46 | Golga4 | 7778 | 0.111 | 0.3520 | No |
| 47 | Clta | 7806 | 0.110 | 0.3563 | No |
| 48 | Rer1 | 7834 | 0.109 | 0.3605 | No |
| 49 | Arfgef1 | 7942 | 0.105 | 0.3611 | No |
| 50 | Mapk1 | 8258 | 0.095 | 0.3524 | No |
| 51 | Vamp4 | 8513 | 0.086 | 0.3458 | No |
| 52 | Tmed10 | 8573 | 0.084 | 0.3474 | No |
| 53 | Sod1 | 8810 | 0.076 | 0.3412 | No |
| 54 | Ap2m1 | 8943 | 0.072 | 0.3391 | No |
| 55 | Pam | 9374 | 0.058 | 0.3237 | No |
| 56 | Ykt6 | 9627 | 0.050 | 0.3154 | No |
| 57 | Stam | 9953 | 0.040 | 0.3036 | No |
| 58 | Cope | 10155 | 0.034 | 0.2967 | No |
| 59 | Atp7a | 10183 | 0.034 | 0.2972 | No |
| 60 | Rab2a | 10547 | 0.023 | 0.2829 | No |
| 61 | Cog2 | 10610 | 0.021 | 0.2813 | No |
| 62 | Yipf6 | 10792 | 0.015 | 0.2744 | No |
| 63 | Vps4b | 11067 | 0.006 | 0.2630 | No |
| 64 | Sec22b | 11638 | -0.010 | 0.2393 | No |
| 65 | Clcn3 | 11682 | -0.012 | 0.2380 | No |
| 66 | Stx7 | 11784 | -0.015 | 0.2345 | No |
| 67 | Gbf1 | 11857 | -0.017 | 0.2322 | No |
| 68 | Cav2 | 12060 | -0.023 | 0.2248 | No |
| 69 | Egfr | 12324 | -0.031 | 0.2151 | No |
| 70 | Sec24d | 12440 | -0.035 | 0.2119 | No |
| 71 | Ergic3 | 12500 | -0.036 | 0.2112 | No |
| 72 | Vamp3 | 12513 | -0.037 | 0.2125 | No |
| 73 | Rab14 | 12523 | -0.037 | 0.2140 | No |
| 74 | Vps45 | 13450 | -0.070 | 0.1780 | No |
| 75 | Rab9 | 13461 | -0.070 | 0.1811 | No |
| 76 | Scamp3 | 13802 | -0.081 | 0.1706 | No |
| 77 | Gosr2 | 13819 | -0.082 | 0.1739 | No |
| 78 | Napg | 14576 | -0.104 | 0.1469 | No |
| 79 | Tspan8 | 15376 | -0.132 | 0.1195 | No |
| 80 | Arfgap3 | 15407 | -0.133 | 0.1248 | No |
| 81 | Rab22a | 15691 | -0.144 | 0.1198 | No |
| 82 | Atp6v1b1 | 16259 | -0.165 | 0.1039 | No |
| 83 | Ap3s1 | 16629 | -0.178 | 0.0969 | No |
| 84 | Dop1a | 17323 | -0.205 | 0.0776 | No |
| 85 | Ica1 | 17994 | -0.234 | 0.0606 | No |
| 86 | Sspn | 18072 | -0.237 | 0.0691 | No |
| 87 | Gnas | 19036 | -0.277 | 0.0418 | No |
| 88 | Tsg101 | 19187 | -0.285 | 0.0494 | No |
| 89 | Ocrl | 19613 | -0.306 | 0.0464 | No |
| 90 | Sgms1 | 19883 | -0.320 | 0.0508 | No |
| 91 | Bet1 | 20708 | -0.371 | 0.0340 | No |
| 92 | Krt18 | 22772 | -0.580 | -0.0251 | No |
| 93 | Cd63 | 22806 | -0.586 | 0.0023 | No |
| 94 | Sh3gl2 | 23123 | -0.658 | 0.0213 | No |