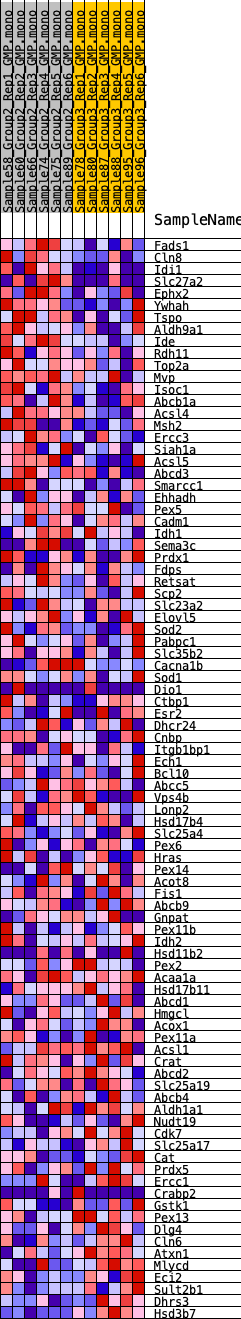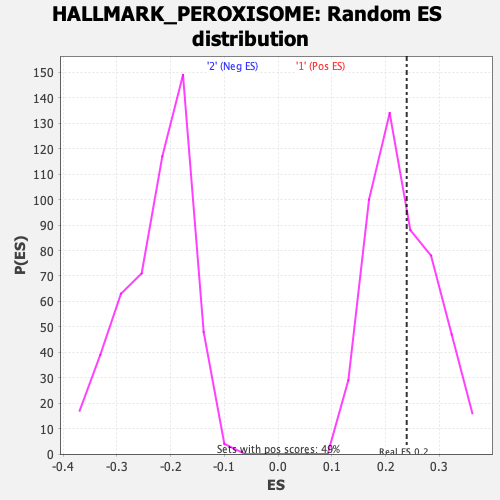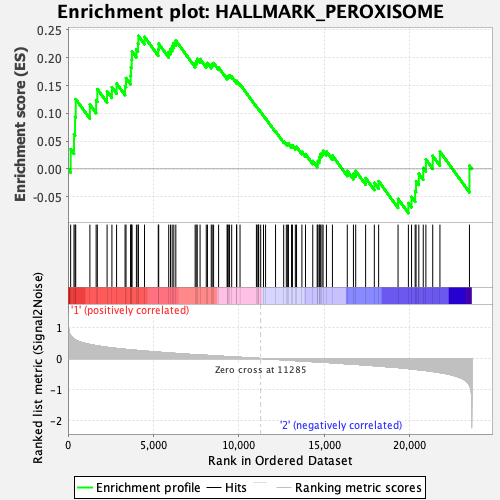
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group2_versus_Group3.GMP.mono_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | GMP.mono_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | 0.23931447 |
| Normalized Enrichment Score (NES) | 1.0467347 |
| Nominal p-value | 0.37398374 |
| FDR q-value | 0.8442202 |
| FWER p-Value | 0.99 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Fads1 | 155 | 0.740 | 0.0353 | Yes |
| 2 | Cln8 | 350 | 0.624 | 0.0623 | Yes |
| 3 | Idi1 | 414 | 0.603 | 0.0937 | Yes |
| 4 | Slc27a2 | 452 | 0.590 | 0.1255 | Yes |
| 5 | Ephx2 | 1281 | 0.450 | 0.1158 | Yes |
| 6 | Ywhah | 1646 | 0.411 | 0.1236 | Yes |
| 7 | Tspo | 1716 | 0.406 | 0.1436 | Yes |
| 8 | Aldh9a1 | 2287 | 0.358 | 0.1396 | Yes |
| 9 | Ide | 2569 | 0.342 | 0.1470 | Yes |
| 10 | Rdh11 | 2842 | 0.322 | 0.1537 | Yes |
| 11 | Top2a | 3338 | 0.295 | 0.1493 | Yes |
| 12 | Mvp | 3394 | 0.292 | 0.1634 | Yes |
| 13 | Isoc1 | 3663 | 0.279 | 0.1678 | Yes |
| 14 | Abcb1a | 3681 | 0.278 | 0.1829 | Yes |
| 15 | Acsl4 | 3726 | 0.275 | 0.1966 | Yes |
| 16 | Msh2 | 3743 | 0.275 | 0.2114 | Yes |
| 17 | Ercc3 | 4002 | 0.261 | 0.2152 | Yes |
| 18 | Siah1a | 4095 | 0.257 | 0.2258 | Yes |
| 19 | Acsl5 | 4119 | 0.256 | 0.2393 | Yes |
| 20 | Abcd3 | 4476 | 0.237 | 0.2376 | No |
| 21 | Smarcc1 | 5280 | 0.202 | 0.2149 | No |
| 22 | Ehhadh | 5306 | 0.201 | 0.2252 | No |
| 23 | Pex5 | 5892 | 0.178 | 0.2104 | No |
| 24 | Cadm1 | 5995 | 0.174 | 0.2159 | No |
| 25 | Idh1 | 6094 | 0.170 | 0.2213 | No |
| 26 | Sema3c | 6191 | 0.166 | 0.2266 | No |
| 27 | Prdx1 | 6308 | 0.162 | 0.2309 | No |
| 28 | Fdps | 7438 | 0.123 | 0.1898 | No |
| 29 | Retsat | 7499 | 0.121 | 0.1941 | No |
| 30 | Scp2 | 7561 | 0.118 | 0.1982 | No |
| 31 | Slc23a2 | 7726 | 0.113 | 0.1976 | No |
| 32 | Elovl5 | 8088 | 0.101 | 0.1879 | No |
| 33 | Sod2 | 8158 | 0.098 | 0.1906 | No |
| 34 | Pabpc1 | 8380 | 0.091 | 0.1863 | No |
| 35 | Slc35b2 | 8436 | 0.089 | 0.1890 | No |
| 36 | Cacna1b | 8518 | 0.086 | 0.1904 | No |
| 37 | Sod1 | 8810 | 0.076 | 0.1824 | No |
| 38 | Dio1 | 9307 | 0.060 | 0.1647 | No |
| 39 | Ctbp1 | 9338 | 0.059 | 0.1667 | No |
| 40 | Esr2 | 9405 | 0.057 | 0.1671 | No |
| 41 | Dhcr24 | 9452 | 0.055 | 0.1683 | No |
| 42 | Cnbp | 9576 | 0.051 | 0.1660 | No |
| 43 | Itgb1bp1 | 9860 | 0.042 | 0.1563 | No |
| 44 | Ech1 | 9872 | 0.042 | 0.1583 | No |
| 45 | Bcl10 | 10064 | 0.037 | 0.1522 | No |
| 46 | Abcc5 | 11022 | 0.007 | 0.1120 | No |
| 47 | Vps4b | 11067 | 0.006 | 0.1105 | No |
| 48 | Lonp2 | 11146 | 0.004 | 0.1074 | No |
| 49 | Hsd17b4 | 11155 | 0.003 | 0.1072 | No |
| 50 | Slc25a4 | 11273 | 0.000 | 0.1023 | No |
| 51 | Pex6 | 11437 | -0.003 | 0.0955 | No |
| 52 | Hras | 11558 | -0.007 | 0.0908 | No |
| 53 | Pex14 | 12142 | -0.025 | 0.0674 | No |
| 54 | Acot8 | 12618 | -0.040 | 0.0495 | No |
| 55 | Fis1 | 12772 | -0.045 | 0.0456 | No |
| 56 | Abcb9 | 12850 | -0.048 | 0.0450 | No |
| 57 | Gnpat | 12896 | -0.050 | 0.0459 | No |
| 58 | Pex11b | 13078 | -0.056 | 0.0413 | No |
| 59 | Idh2 | 13130 | -0.058 | 0.0424 | No |
| 60 | Hsd11b2 | 13300 | -0.064 | 0.0389 | No |
| 61 | Pex2 | 13370 | -0.066 | 0.0397 | No |
| 62 | Acaa1a | 13684 | -0.077 | 0.0308 | No |
| 63 | Hsd17b11 | 13895 | -0.084 | 0.0266 | No |
| 64 | Abcd1 | 14319 | -0.098 | 0.0142 | No |
| 65 | Hmgcl | 14577 | -0.105 | 0.0092 | No |
| 66 | Acox1 | 14612 | -0.106 | 0.0137 | No |
| 67 | Pex11a | 14689 | -0.108 | 0.0166 | No |
| 68 | Acsl1 | 14727 | -0.110 | 0.0213 | No |
| 69 | Crat | 14761 | -0.112 | 0.0262 | No |
| 70 | Abcd2 | 14856 | -0.115 | 0.0287 | No |
| 71 | Slc25a19 | 14923 | -0.118 | 0.0325 | No |
| 72 | Abcb4 | 15119 | -0.124 | 0.0312 | No |
| 73 | Aldh1a1 | 15470 | -0.135 | 0.0240 | No |
| 74 | Nudt19 | 16339 | -0.168 | -0.0034 | No |
| 75 | Cdk7 | 16699 | -0.180 | -0.0085 | No |
| 76 | Slc25a17 | 16831 | -0.184 | -0.0036 | No |
| 77 | Cat | 17412 | -0.209 | -0.0165 | No |
| 78 | Prdx5 | 17921 | -0.231 | -0.0250 | No |
| 79 | Ercc1 | 18173 | -0.241 | -0.0221 | No |
| 80 | Crabp2 | 19309 | -0.289 | -0.0539 | No |
| 81 | Gstk1 | 19914 | -0.321 | -0.0614 | No |
| 82 | Pex13 | 20098 | -0.334 | -0.0503 | No |
| 83 | Dlg4 | 20309 | -0.346 | -0.0396 | No |
| 84 | Cln6 | 20362 | -0.350 | -0.0221 | No |
| 85 | Atxn1 | 20524 | -0.360 | -0.0085 | No |
| 86 | Mlycd | 20783 | -0.375 | 0.0017 | No |
| 87 | Eci2 | 20937 | -0.386 | 0.0170 | No |
| 88 | Sult2b1 | 21337 | -0.414 | 0.0235 | No |
| 89 | Dhrs3 | 21759 | -0.451 | 0.0311 | No |
| 90 | Hsd3b7 | 23486 | -0.851 | 0.0059 | No |