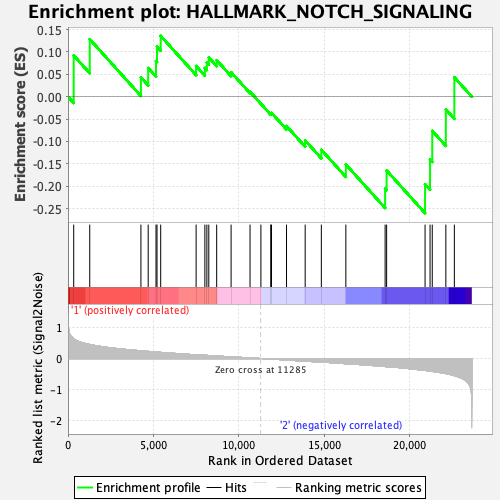
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group2_versus_Group3.GMP.mono_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | GMP.mono_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_NOTCH_SIGNALING |
| Enrichment Score (ES) | -0.26032513 |
| Normalized Enrichment Score (NES) | -0.7895775 |
| Nominal p-value | 0.7191235 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
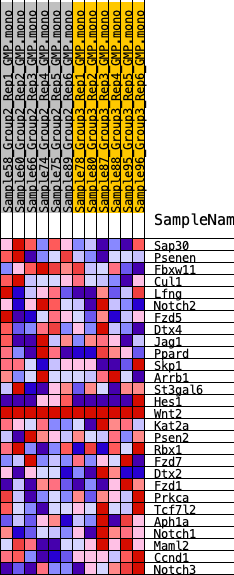
| 1 | Sap30 | 334 | 0.630 | 0.0921 | No |
| 2 | Psenen | 1272 | 0.451 | 0.1285 | No |
| 3 | Fbxw11 | 4266 | 0.248 | 0.0434 | No |
| 4 | Cul1 | 4691 | 0.229 | 0.0640 | No |
| 5 | Lfng | 5149 | 0.208 | 0.0797 | No |
| 6 | Notch2 | 5207 | 0.205 | 0.1119 | No |
| 7 | Fzd5 | 5423 | 0.197 | 0.1359 | No |
| 8 | Dtx4 | 7495 | 0.121 | 0.0686 | No |
| 9 | Jag1 | 8007 | 0.103 | 0.0643 | No |
| 10 | Ppard | 8124 | 0.099 | 0.0762 | No |
| 11 | Skp1 | 8237 | 0.095 | 0.0875 | No |
| 12 | Arrb1 | 8698 | 0.080 | 0.0815 | No |
| 13 | St3gal6 | 9541 | 0.052 | 0.0547 | No |
| 14 | Hes1 | 10650 | 0.019 | 0.0110 | No |
| 15 | Wnt2 | 11285 | 0.000 | -0.0159 | No |
| 16 | Kat2a | 11860 | -0.017 | -0.0373 | No |
| 17 | Psen2 | 11906 | -0.018 | -0.0362 | No |
| 18 | Rbx1 | 12784 | -0.045 | -0.0657 | No |
| 19 | Fzd7 | 13875 | -0.084 | -0.0978 | No |
| 20 | Dtx2 | 14821 | -0.114 | -0.1186 | No |
| 21 | Fzd1 | 16255 | -0.165 | -0.1516 | No |
| 22 | Prkca | 18552 | -0.257 | -0.2054 | Yes |
| 23 | Tcf7l2 | 18640 | -0.261 | -0.1651 | Yes |
| 24 | Aph1a | 20889 | -0.382 | -0.1958 | Yes |
| 25 | Notch1 | 21181 | -0.402 | -0.1404 | Yes |
| 26 | Maml2 | 21310 | -0.412 | -0.0764 | Yes |
| 27 | Ccnd1 | 22105 | -0.483 | -0.0285 | Yes |
| 28 | Notch3 | 22602 | -0.550 | 0.0433 | Yes |