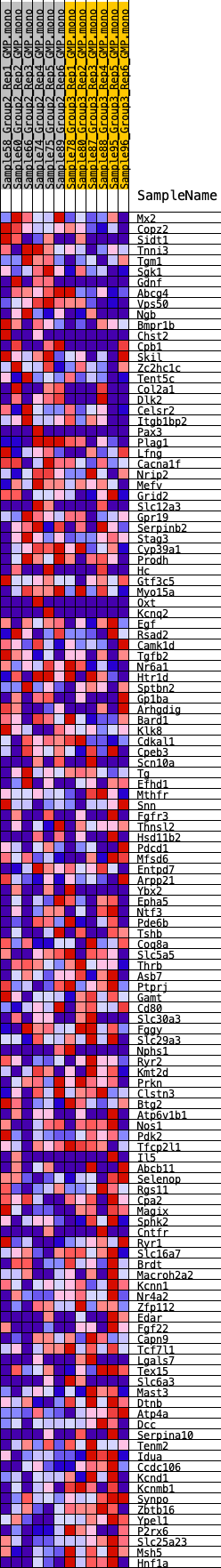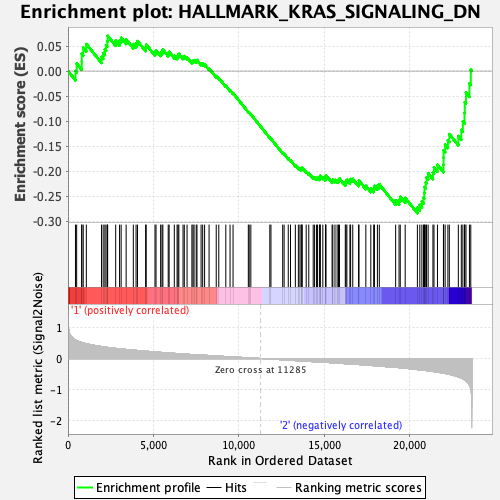
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group2_versus_Group3.GMP.mono_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | GMP.mono_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | -0.28385764 |
| Normalized Enrichment Score (NES) | -1.0990208 |
| Nominal p-value | 0.27920792 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.967 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Mx2 | 437 | 0.594 | -0.0000 | No |
| 2 | Copz2 | 507 | 0.576 | 0.0151 | No |
| 3 | Sidt1 | 803 | 0.515 | 0.0187 | No |
| 4 | Tnni3 | 805 | 0.515 | 0.0348 | No |
| 5 | Tgm1 | 890 | 0.499 | 0.0468 | No |
| 6 | Sgk1 | 1072 | 0.473 | 0.0539 | No |
| 7 | Gdnf | 1961 | 0.385 | 0.0282 | No |
| 8 | Abcg4 | 2057 | 0.379 | 0.0360 | No |
| 9 | Vps50 | 2157 | 0.370 | 0.0434 | No |
| 10 | Ngb | 2226 | 0.365 | 0.0519 | No |
| 11 | Bmpr1b | 2305 | 0.357 | 0.0597 | No |
| 12 | Chst2 | 2321 | 0.356 | 0.0703 | No |
| 13 | Cpb1 | 2790 | 0.327 | 0.0606 | No |
| 14 | Skil | 3017 | 0.311 | 0.0607 | No |
| 15 | Zc2hc1c | 3109 | 0.307 | 0.0664 | No |
| 16 | Tent5c | 3401 | 0.291 | 0.0632 | No |
| 17 | Col2a1 | 3821 | 0.271 | 0.0538 | No |
| 18 | Dlk2 | 3982 | 0.263 | 0.0553 | No |
| 19 | Celsr2 | 4065 | 0.258 | 0.0598 | No |
| 20 | Itgb1bp2 | 4541 | 0.233 | 0.0469 | No |
| 21 | Pax3 | 4575 | 0.231 | 0.0528 | No |
| 22 | Plag1 | 5089 | 0.211 | 0.0375 | No |
| 23 | Lfng | 5149 | 0.208 | 0.0415 | No |
| 24 | Cacna1f | 5419 | 0.197 | 0.0362 | No |
| 25 | Nrip2 | 5499 | 0.193 | 0.0389 | No |
| 26 | Mefv | 5550 | 0.191 | 0.0428 | No |
| 27 | Grid2 | 5863 | 0.178 | 0.0351 | No |
| 28 | Slc12a3 | 5916 | 0.177 | 0.0384 | No |
| 29 | Gpr19 | 6219 | 0.165 | 0.0307 | No |
| 30 | Serpinb2 | 6382 | 0.159 | 0.0288 | No |
| 31 | Stag3 | 6436 | 0.157 | 0.0315 | No |
| 32 | Cyp39a1 | 6485 | 0.155 | 0.0343 | No |
| 33 | Prodh | 6730 | 0.146 | 0.0285 | No |
| 34 | Hc | 6804 | 0.143 | 0.0298 | No |
| 35 | Gtf3c5 | 6965 | 0.137 | 0.0273 | No |
| 36 | Myo15a | 7248 | 0.126 | 0.0193 | No |
| 37 | Oxt | 7312 | 0.124 | 0.0205 | No |
| 38 | Kcnq2 | 7375 | 0.123 | 0.0217 | No |
| 39 | Egf | 7500 | 0.121 | 0.0202 | No |
| 40 | Rsad2 | 7545 | 0.119 | 0.0220 | No |
| 41 | Camk1d | 7786 | 0.111 | 0.0153 | No |
| 42 | Tgfb2 | 7879 | 0.107 | 0.0147 | No |
| 43 | Nr6a1 | 7992 | 0.104 | 0.0132 | No |
| 44 | Htr1d | 8253 | 0.095 | 0.0051 | No |
| 45 | Sptbn2 | 8672 | 0.081 | -0.0101 | No |
| 46 | Gp1ba | 8821 | 0.076 | -0.0141 | No |
| 47 | Arhgdig | 9232 | 0.062 | -0.0295 | No |
| 48 | Bard1 | 9480 | 0.054 | -0.0384 | No |
| 49 | Klk8 | 9656 | 0.049 | -0.0443 | No |
| 50 | Cdkal1 | 10549 | 0.023 | -0.0815 | No |
| 51 | Cpeb3 | 10599 | 0.021 | -0.0830 | No |
| 52 | Scn10a | 10696 | 0.018 | -0.0865 | No |
| 53 | Tg | 11807 | -0.016 | -0.1332 | No |
| 54 | Efhd1 | 11873 | -0.017 | -0.1354 | No |
| 55 | Mthfr | 12560 | -0.038 | -0.1635 | No |
| 56 | Snn | 12643 | -0.041 | -0.1657 | No |
| 57 | Fgfr3 | 12886 | -0.049 | -0.1744 | No |
| 58 | Thnsl2 | 13017 | -0.054 | -0.1783 | No |
| 59 | Hsd11b2 | 13300 | -0.064 | -0.1883 | No |
| 60 | Pdcd1 | 13477 | -0.070 | -0.1935 | No |
| 61 | Mfsd6 | 13569 | -0.073 | -0.1951 | No |
| 62 | Entpd7 | 13645 | -0.076 | -0.1959 | No |
| 63 | Arpp21 | 13687 | -0.078 | -0.1952 | No |
| 64 | Ybx2 | 13695 | -0.078 | -0.1931 | No |
| 65 | Epha5 | 13935 | -0.085 | -0.2006 | No |
| 66 | Ntf3 | 14086 | -0.090 | -0.2042 | No |
| 67 | Pde6b | 14345 | -0.099 | -0.2121 | No |
| 68 | Tshb | 14426 | -0.101 | -0.2123 | No |
| 69 | Coq8a | 14540 | -0.103 | -0.2139 | No |
| 70 | Slc5a5 | 14593 | -0.105 | -0.2128 | No |
| 71 | Thrb | 14707 | -0.109 | -0.2142 | No |
| 72 | Asb7 | 14751 | -0.111 | -0.2125 | No |
| 73 | Ptprj | 14753 | -0.111 | -0.2091 | No |
| 74 | Gamt | 14908 | -0.117 | -0.2120 | No |
| 75 | Cd80 | 15062 | -0.122 | -0.2147 | No |
| 76 | Slc30a3 | 15067 | -0.122 | -0.2110 | No |
| 77 | Fggy | 15094 | -0.123 | -0.2083 | No |
| 78 | Slc29a3 | 15450 | -0.135 | -0.2192 | No |
| 79 | Nphs1 | 15493 | -0.137 | -0.2167 | No |
| 80 | Ryr2 | 15619 | -0.141 | -0.2176 | No |
| 81 | Kmt2d | 15745 | -0.146 | -0.2184 | No |
| 82 | Prkn | 15826 | -0.149 | -0.2171 | No |
| 83 | Clstn3 | 15883 | -0.152 | -0.2147 | No |
| 84 | Btg2 | 16222 | -0.164 | -0.2240 | No |
| 85 | Atp6v1b1 | 16259 | -0.165 | -0.2203 | No |
| 86 | Nos1 | 16315 | -0.167 | -0.2175 | No |
| 87 | Pdk2 | 16493 | -0.172 | -0.2196 | No |
| 88 | Tfcp2l1 | 16533 | -0.174 | -0.2158 | No |
| 89 | Il5 | 16659 | -0.179 | -0.2155 | No |
| 90 | Abcb11 | 17001 | -0.192 | -0.2240 | No |
| 91 | Selenop | 17022 | -0.193 | -0.2188 | No |
| 92 | Rgs11 | 17424 | -0.210 | -0.2293 | No |
| 93 | Cpa2 | 17715 | -0.221 | -0.2348 | No |
| 94 | Magix | 17884 | -0.228 | -0.2348 | No |
| 95 | Sphk2 | 17927 | -0.231 | -0.2293 | No |
| 96 | Cntfr | 18101 | -0.238 | -0.2292 | No |
| 97 | Ryr1 | 18212 | -0.242 | -0.2263 | No |
| 98 | Slc16a7 | 19169 | -0.284 | -0.2581 | No |
| 99 | Brdt | 19367 | -0.292 | -0.2574 | No |
| 100 | Macroh2a2 | 19438 | -0.296 | -0.2511 | No |
| 101 | Kcnn1 | 19726 | -0.313 | -0.2535 | No |
| 102 | Nr4a2 | 20440 | -0.355 | -0.2727 | Yes |
| 103 | Zfp112 | 20582 | -0.364 | -0.2674 | Yes |
| 104 | Edar | 20703 | -0.371 | -0.2608 | Yes |
| 105 | Fgf22 | 20800 | -0.377 | -0.2531 | Yes |
| 106 | Capn9 | 20844 | -0.379 | -0.2431 | Yes |
| 107 | Tcf7l1 | 20859 | -0.380 | -0.2318 | Yes |
| 108 | Lgals7 | 20927 | -0.385 | -0.2226 | Yes |
| 109 | Tex15 | 20975 | -0.387 | -0.2124 | Yes |
| 110 | Slc6a3 | 21068 | -0.394 | -0.2040 | Yes |
| 111 | Mast3 | 21350 | -0.415 | -0.2030 | Yes |
| 112 | Dtnb | 21411 | -0.422 | -0.1923 | Yes |
| 113 | Atp4a | 21614 | -0.438 | -0.1872 | Yes |
| 114 | Dcc | 21964 | -0.469 | -0.1874 | Yes |
| 115 | Serpina10 | 21969 | -0.469 | -0.1729 | Yes |
| 116 | Tenm2 | 21973 | -0.469 | -0.1583 | Yes |
| 117 | Idua | 22064 | -0.479 | -0.1471 | Yes |
| 118 | Ccdc106 | 22225 | -0.497 | -0.1384 | Yes |
| 119 | Kcnd1 | 22308 | -0.506 | -0.1260 | Yes |
| 120 | Kcnmb1 | 22835 | -0.594 | -0.1298 | Yes |
| 121 | Synpo | 23009 | -0.627 | -0.1175 | Yes |
| 122 | Zbtb16 | 23100 | -0.652 | -0.1009 | Yes |
| 123 | Ypel1 | 23197 | -0.686 | -0.0835 | Yes |
| 124 | P2rx6 | 23213 | -0.691 | -0.0625 | Yes |
| 125 | Slc25a23 | 23283 | -0.722 | -0.0428 | Yes |
| 126 | Msh5 | 23485 | -0.851 | -0.0247 | Yes |
| 127 | Hnf1a | 23564 | -0.977 | 0.0026 | Yes |