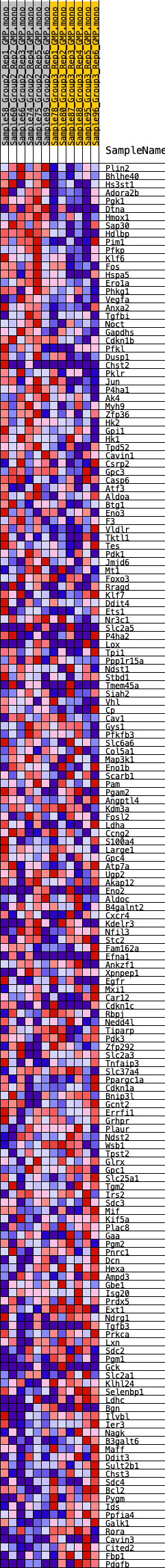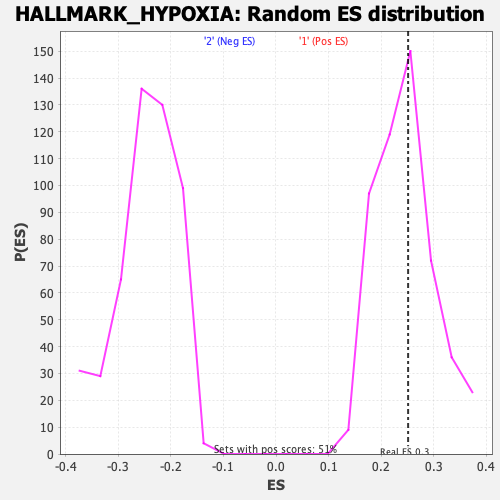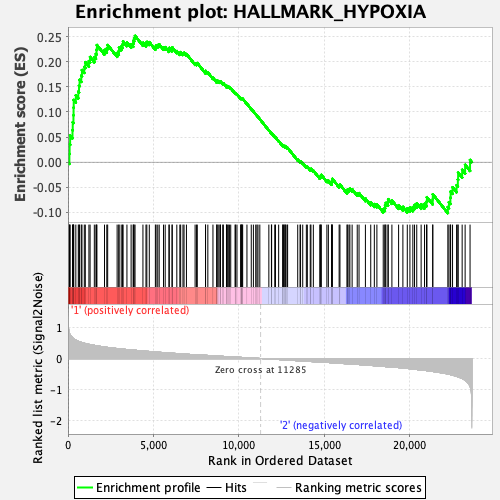
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group2_versus_Group3.GMP.mono_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | GMP.mono_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_HYPOXIA |
| Enrichment Score (ES) | 0.25160852 |
| Normalized Enrichment Score (NES) | 1.0245882 |
| Nominal p-value | 0.42885375 |
| FDR q-value | 0.8142715 |
| FWER p-Value | 0.994 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Plin2 | 81 | 0.821 | 0.0167 | Yes |
| 2 | Bhlhe40 | 95 | 0.795 | 0.0357 | Yes |
| 3 | Hs3st1 | 125 | 0.764 | 0.0532 | Yes |
| 4 | Adora2b | 263 | 0.667 | 0.0638 | Yes |
| 5 | Pgk1 | 277 | 0.662 | 0.0795 | Yes |
| 6 | Dtna | 319 | 0.640 | 0.0934 | Yes |
| 7 | Hmox1 | 326 | 0.634 | 0.1088 | Yes |
| 8 | Sap30 | 334 | 0.630 | 0.1239 | Yes |
| 9 | Hdlbp | 458 | 0.589 | 0.1332 | Yes |
| 10 | Pim1 | 602 | 0.554 | 0.1407 | Yes |
| 11 | Pfkp | 645 | 0.543 | 0.1522 | Yes |
| 12 | Klf6 | 678 | 0.536 | 0.1640 | Yes |
| 13 | Fos | 777 | 0.519 | 0.1726 | Yes |
| 14 | Hspa5 | 816 | 0.513 | 0.1835 | Yes |
| 15 | Ero1a | 953 | 0.491 | 0.1898 | Yes |
| 16 | Phkg1 | 1012 | 0.483 | 0.1992 | Yes |
| 17 | Vegfa | 1224 | 0.456 | 0.2014 | Yes |
| 18 | Anxa2 | 1292 | 0.448 | 0.2096 | Yes |
| 19 | Tgfbi | 1540 | 0.424 | 0.2094 | Yes |
| 20 | Noct | 1633 | 0.412 | 0.2157 | Yes |
| 21 | Gapdhs | 1669 | 0.409 | 0.2242 | Yes |
| 22 | Cdkn1b | 1691 | 0.408 | 0.2333 | Yes |
| 23 | Pfkl | 2136 | 0.371 | 0.2235 | Yes |
| 24 | Dusp1 | 2270 | 0.360 | 0.2267 | Yes |
| 25 | Chst2 | 2321 | 0.356 | 0.2333 | Yes |
| 26 | Pklr | 2874 | 0.320 | 0.2176 | Yes |
| 27 | Jun | 2972 | 0.313 | 0.2212 | Yes |
| 28 | P4ha1 | 2987 | 0.313 | 0.2283 | Yes |
| 29 | Ak4 | 3113 | 0.307 | 0.2305 | Yes |
| 30 | Myh9 | 3191 | 0.303 | 0.2347 | Yes |
| 31 | Zfp36 | 3222 | 0.302 | 0.2408 | Yes |
| 32 | Hk2 | 3448 | 0.290 | 0.2383 | Yes |
| 33 | Gpi1 | 3692 | 0.278 | 0.2348 | Yes |
| 34 | Hk1 | 3812 | 0.272 | 0.2364 | Yes |
| 35 | Tpd52 | 3829 | 0.270 | 0.2423 | Yes |
| 36 | Cavin1 | 3870 | 0.268 | 0.2472 | Yes |
| 37 | Csrp2 | 3921 | 0.266 | 0.2516 | Yes |
| 38 | Gpc3 | 4384 | 0.241 | 0.2378 | No |
| 39 | Casp6 | 4562 | 0.232 | 0.2360 | No |
| 40 | Atf3 | 4612 | 0.230 | 0.2396 | No |
| 41 | Aldoa | 4756 | 0.227 | 0.2390 | No |
| 42 | Btg1 | 5111 | 0.210 | 0.2291 | No |
| 43 | Eno3 | 5152 | 0.208 | 0.2325 | No |
| 44 | F3 | 5251 | 0.203 | 0.2333 | No |
| 45 | Vldlr | 5335 | 0.201 | 0.2347 | No |
| 46 | Tktl1 | 5588 | 0.189 | 0.2286 | No |
| 47 | Tes | 5686 | 0.186 | 0.2291 | No |
| 48 | Pdk1 | 5907 | 0.177 | 0.2240 | No |
| 49 | Jmjd6 | 5923 | 0.176 | 0.2277 | No |
| 50 | Mt1 | 6086 | 0.170 | 0.2250 | No |
| 51 | Foxo3 | 6099 | 0.170 | 0.2286 | No |
| 52 | Rragd | 6366 | 0.160 | 0.2212 | No |
| 53 | Klf7 | 6543 | 0.153 | 0.2175 | No |
| 54 | Ddit4 | 6591 | 0.151 | 0.2192 | No |
| 55 | Ets1 | 6726 | 0.146 | 0.2171 | No |
| 56 | Nr3c1 | 6788 | 0.143 | 0.2180 | No |
| 57 | Slc2a5 | 6929 | 0.139 | 0.2154 | No |
| 58 | P4ha2 | 7436 | 0.123 | 0.1969 | No |
| 59 | Lox | 7521 | 0.120 | 0.1962 | No |
| 60 | Tpi1 | 7563 | 0.118 | 0.1974 | No |
| 61 | Ppp1r15a | 8045 | 0.102 | 0.1794 | No |
| 62 | Ndst1 | 8047 | 0.102 | 0.1818 | No |
| 63 | Stbd1 | 8181 | 0.098 | 0.1786 | No |
| 64 | Tmem45a | 8480 | 0.087 | 0.1680 | No |
| 65 | Siah2 | 8703 | 0.080 | 0.1605 | No |
| 66 | Vhl | 8705 | 0.080 | 0.1624 | No |
| 67 | Cp | 8767 | 0.078 | 0.1617 | No |
| 68 | Cav1 | 8832 | 0.076 | 0.1609 | No |
| 69 | Gys1 | 8899 | 0.074 | 0.1599 | No |
| 70 | Pfkfb3 | 8921 | 0.073 | 0.1607 | No |
| 71 | Slc6a6 | 9053 | 0.069 | 0.1568 | No |
| 72 | Col5a1 | 9088 | 0.067 | 0.1570 | No |
| 73 | Map3k1 | 9270 | 0.061 | 0.1508 | No |
| 74 | Eno1b | 9276 | 0.061 | 0.1521 | No |
| 75 | Scarb1 | 9343 | 0.059 | 0.1507 | No |
| 76 | Pam | 9374 | 0.058 | 0.1509 | No |
| 77 | Pgam2 | 9443 | 0.056 | 0.1494 | No |
| 78 | Angptl4 | 9520 | 0.053 | 0.1474 | No |
| 79 | Kdm3a | 9773 | 0.045 | 0.1378 | No |
| 80 | Fosl2 | 9805 | 0.044 | 0.1375 | No |
| 81 | Ldha | 9885 | 0.042 | 0.1352 | No |
| 82 | Ccng2 | 10102 | 0.036 | 0.1269 | No |
| 83 | S100a4 | 10125 | 0.035 | 0.1268 | No |
| 84 | Large1 | 10136 | 0.035 | 0.1272 | No |
| 85 | Gpc4 | 10173 | 0.034 | 0.1265 | No |
| 86 | Atp7a | 10183 | 0.034 | 0.1270 | No |
| 87 | Ugp2 | 10218 | 0.032 | 0.1263 | No |
| 88 | Akap12 | 10469 | 0.025 | 0.1163 | No |
| 89 | Eno2 | 10721 | 0.017 | 0.1060 | No |
| 90 | Aldoc | 10846 | 0.013 | 0.1010 | No |
| 91 | B4galnt2 | 10983 | 0.009 | 0.0954 | No |
| 92 | Cxcr4 | 11053 | 0.007 | 0.0927 | No |
| 93 | Kdelr3 | 11114 | 0.005 | 0.0902 | No |
| 94 | Nfil3 | 11222 | 0.002 | 0.0857 | No |
| 95 | Stc2 | 11750 | -0.014 | 0.0636 | No |
| 96 | Fam162a | 11901 | -0.018 | 0.0576 | No |
| 97 | Efna1 | 11913 | -0.018 | 0.0576 | No |
| 98 | Ankzf1 | 12096 | -0.024 | 0.0504 | No |
| 99 | Xpnpep1 | 12138 | -0.025 | 0.0493 | No |
| 100 | Egfr | 12324 | -0.031 | 0.0422 | No |
| 101 | Mxi1 | 12562 | -0.038 | 0.0330 | No |
| 102 | Car12 | 12574 | -0.038 | 0.0334 | No |
| 103 | Cdkn1c | 12613 | -0.039 | 0.0328 | No |
| 104 | Rbpj | 12693 | -0.042 | 0.0305 | No |
| 105 | Nedd4l | 12695 | -0.042 | 0.0315 | No |
| 106 | Tiparp | 12709 | -0.043 | 0.0320 | No |
| 107 | Pdk3 | 12807 | -0.046 | 0.0290 | No |
| 108 | Zfp292 | 12848 | -0.048 | 0.0284 | No |
| 109 | Slc2a3 | 13439 | -0.069 | 0.0050 | No |
| 110 | Tnfaip3 | 13575 | -0.074 | 0.0010 | No |
| 111 | Slc37a4 | 13605 | -0.074 | 0.0016 | No |
| 112 | Ppargc1a | 13732 | -0.079 | -0.0018 | No |
| 113 | Cdkn1a | 13944 | -0.086 | -0.0087 | No |
| 114 | Bnip3l | 14002 | -0.087 | -0.0090 | No |
| 115 | Gcnt2 | 14171 | -0.093 | -0.0139 | No |
| 116 | Errfi1 | 14212 | -0.094 | -0.0133 | No |
| 117 | Grhpr | 14353 | -0.099 | -0.0168 | No |
| 118 | Plaur | 14731 | -0.110 | -0.0302 | No |
| 119 | Ndst2 | 14764 | -0.112 | -0.0288 | No |
| 120 | Wsb1 | 14797 | -0.113 | -0.0274 | No |
| 121 | Tpst2 | 14815 | -0.113 | -0.0253 | No |
| 122 | Glrx | 15145 | -0.125 | -0.0363 | No |
| 123 | Gpc1 | 15232 | -0.127 | -0.0368 | No |
| 124 | Slc25a1 | 15425 | -0.134 | -0.0417 | No |
| 125 | Tgm2 | 15427 | -0.134 | -0.0385 | No |
| 126 | Irs2 | 15454 | -0.135 | -0.0363 | No |
| 127 | Sdc3 | 15461 | -0.135 | -0.0332 | No |
| 128 | Mif | 15862 | -0.151 | -0.0465 | No |
| 129 | Kif5a | 15910 | -0.153 | -0.0448 | No |
| 130 | Plac8 | 16325 | -0.167 | -0.0583 | No |
| 131 | Gaa | 16340 | -0.168 | -0.0548 | No |
| 132 | Pgm2 | 16435 | -0.171 | -0.0546 | No |
| 133 | Pnrc1 | 16490 | -0.172 | -0.0527 | No |
| 134 | Dcn | 16622 | -0.177 | -0.0539 | No |
| 135 | Hexa | 16921 | -0.188 | -0.0620 | No |
| 136 | Ampd3 | 17026 | -0.193 | -0.0617 | No |
| 137 | Gbe1 | 17399 | -0.208 | -0.0725 | No |
| 138 | Isg20 | 17711 | -0.221 | -0.0803 | No |
| 139 | Prdx5 | 17921 | -0.231 | -0.0835 | No |
| 140 | Ext1 | 18071 | -0.237 | -0.0841 | No |
| 141 | Ndrg1 | 18442 | -0.253 | -0.0936 | No |
| 142 | Tgfb3 | 18535 | -0.257 | -0.0912 | No |
| 143 | Prkca | 18552 | -0.257 | -0.0856 | No |
| 144 | Lxn | 18597 | -0.260 | -0.0811 | No |
| 145 | Sdc2 | 18722 | -0.264 | -0.0799 | No |
| 146 | Pgm1 | 18732 | -0.264 | -0.0738 | No |
| 147 | Gck | 18949 | -0.273 | -0.0763 | No |
| 148 | Slc2a1 | 19341 | -0.291 | -0.0858 | No |
| 149 | Klhl24 | 19592 | -0.305 | -0.0890 | No |
| 150 | Selenbp1 | 19849 | -0.319 | -0.0921 | No |
| 151 | Ldhc | 20003 | -0.327 | -0.0906 | No |
| 152 | Bgn | 20174 | -0.338 | -0.0895 | No |
| 153 | Ilvbl | 20273 | -0.345 | -0.0852 | No |
| 154 | Ier3 | 20403 | -0.353 | -0.0821 | No |
| 155 | Nagk | 20659 | -0.368 | -0.0839 | No |
| 156 | B3galt6 | 20858 | -0.380 | -0.0830 | No |
| 157 | Maff | 20977 | -0.388 | -0.0785 | No |
| 158 | Ddit3 | 20997 | -0.390 | -0.0698 | No |
| 159 | Sult2b1 | 21337 | -0.414 | -0.0740 | No |
| 160 | Chst3 | 21341 | -0.414 | -0.0640 | No |
| 161 | Sdc4 | 22222 | -0.497 | -0.0893 | No |
| 162 | Bcl2 | 22286 | -0.504 | -0.0796 | No |
| 163 | Pygm | 22371 | -0.516 | -0.0706 | No |
| 164 | Ids | 22385 | -0.518 | -0.0584 | No |
| 165 | Ppfia4 | 22497 | -0.535 | -0.0500 | No |
| 166 | Galk1 | 22733 | -0.572 | -0.0459 | No |
| 167 | Rora | 22807 | -0.586 | -0.0347 | No |
| 168 | Cavin3 | 22823 | -0.591 | -0.0208 | No |
| 169 | Cited2 | 23059 | -0.639 | -0.0151 | No |
| 170 | Fbp1 | 23233 | -0.700 | -0.0053 | No |
| 171 | Pdgfb | 23521 | -0.892 | 0.0044 | No |