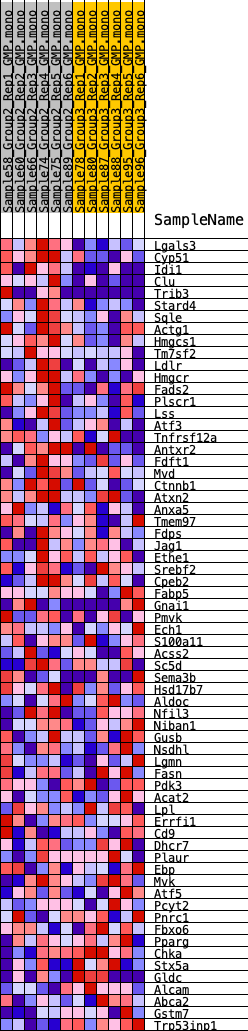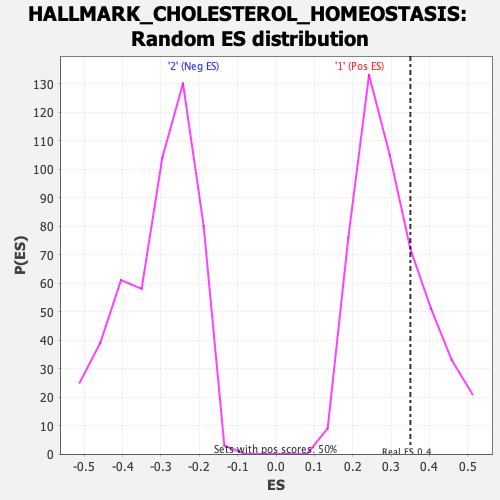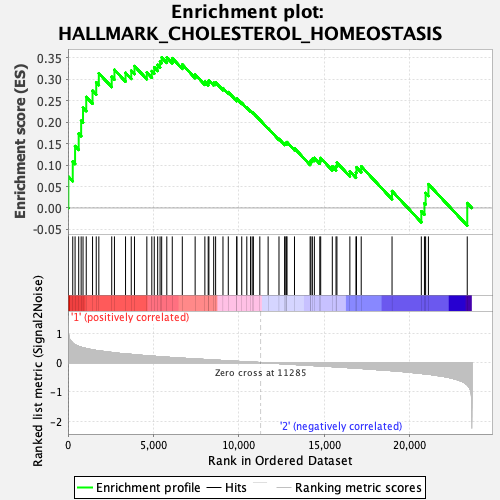
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group2_versus_Group3.GMP.mono_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | GMP.mono_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | 0.3504893 |
| Normalized Enrichment Score (NES) | 1.1581321 |
| Nominal p-value | 0.27 |
| FDR q-value | 0.7984795 |
| FWER p-Value | 0.936 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Lgals3 | 20 | 1.064 | 0.0730 | Yes |
| 2 | Cyp51 | 278 | 0.660 | 0.1080 | Yes |
| 3 | Idi1 | 414 | 0.603 | 0.1441 | Yes |
| 4 | Clu | 626 | 0.546 | 0.1731 | Yes |
| 5 | Trib3 | 769 | 0.520 | 0.2033 | Yes |
| 6 | Stard4 | 879 | 0.502 | 0.2335 | Yes |
| 7 | Sqle | 1067 | 0.473 | 0.2584 | Yes |
| 8 | Actg1 | 1436 | 0.434 | 0.2729 | Yes |
| 9 | Hmgcs1 | 1652 | 0.411 | 0.2923 | Yes |
| 10 | Tm7sf2 | 1804 | 0.397 | 0.3135 | Yes |
| 11 | Ldlr | 2558 | 0.343 | 0.3054 | Yes |
| 12 | Hmgcr | 2717 | 0.332 | 0.3217 | Yes |
| 13 | Fads2 | 3366 | 0.293 | 0.3146 | Yes |
| 14 | Plscr1 | 3700 | 0.277 | 0.3197 | Yes |
| 15 | Lss | 3889 | 0.267 | 0.3303 | Yes |
| 16 | Atf3 | 4612 | 0.230 | 0.3156 | Yes |
| 17 | Tnfrsf12a | 4903 | 0.220 | 0.3186 | Yes |
| 18 | Antxr2 | 5046 | 0.213 | 0.3274 | Yes |
| 19 | Fdft1 | 5246 | 0.204 | 0.3331 | Yes |
| 20 | Mvd | 5386 | 0.199 | 0.3410 | Yes |
| 21 | Ctnnb1 | 5480 | 0.194 | 0.3505 | Yes |
| 22 | Atxn2 | 5780 | 0.182 | 0.3505 | No |
| 23 | Anxa5 | 6100 | 0.170 | 0.3487 | No |
| 24 | Tmem97 | 6687 | 0.147 | 0.3340 | No |
| 25 | Fdps | 7438 | 0.123 | 0.3107 | No |
| 26 | Jag1 | 8007 | 0.103 | 0.2938 | No |
| 27 | Ethe1 | 8202 | 0.097 | 0.2923 | No |
| 28 | Srebf2 | 8243 | 0.095 | 0.2972 | No |
| 29 | Cpeb2 | 8523 | 0.086 | 0.2913 | No |
| 30 | Fabp5 | 8636 | 0.082 | 0.2923 | No |
| 31 | Gnai1 | 9066 | 0.068 | 0.2788 | No |
| 32 | Pmvk | 9377 | 0.058 | 0.2697 | No |
| 33 | Ech1 | 9872 | 0.042 | 0.2516 | No |
| 34 | S100a11 | 9876 | 0.042 | 0.2544 | No |
| 35 | Acss2 | 10164 | 0.034 | 0.2446 | No |
| 36 | Sc5d | 10460 | 0.025 | 0.2338 | No |
| 37 | Sema3b | 10679 | 0.019 | 0.2259 | No |
| 38 | Hsd17b7 | 10774 | 0.015 | 0.2229 | No |
| 39 | Aldoc | 10846 | 0.013 | 0.2208 | No |
| 40 | Nfil3 | 11222 | 0.002 | 0.2050 | No |
| 41 | Niban1 | 11708 | -0.012 | 0.1853 | No |
| 42 | Gusb | 12343 | -0.031 | 0.1606 | No |
| 43 | Nsdhl | 12673 | -0.042 | 0.1495 | No |
| 44 | Lgmn | 12705 | -0.043 | 0.1512 | No |
| 45 | Fasn | 12793 | -0.046 | 0.1507 | No |
| 46 | Pdk3 | 12807 | -0.046 | 0.1533 | No |
| 47 | Acat2 | 13250 | -0.062 | 0.1389 | No |
| 48 | Lpl | 14165 | -0.093 | 0.1066 | No |
| 49 | Errfi1 | 14212 | -0.094 | 0.1111 | No |
| 50 | Cd9 | 14296 | -0.097 | 0.1144 | No |
| 51 | Dhcr7 | 14413 | -0.101 | 0.1165 | No |
| 52 | Plaur | 14731 | -0.110 | 0.1107 | No |
| 53 | Ebp | 14783 | -0.112 | 0.1163 | No |
| 54 | Mvk | 15463 | -0.135 | 0.0969 | No |
| 55 | Atf5 | 15680 | -0.143 | 0.0976 | No |
| 56 | Pcyt2 | 15733 | -0.145 | 0.1055 | No |
| 57 | Pnrc1 | 16490 | -0.172 | 0.0854 | No |
| 58 | Fbxo6 | 16847 | -0.185 | 0.0831 | No |
| 59 | Pparg | 16877 | -0.186 | 0.0948 | No |
| 60 | Chka | 17154 | -0.198 | 0.0969 | No |
| 61 | Stx5a | 18957 | -0.273 | 0.0393 | No |
| 62 | Gldc | 20670 | -0.369 | -0.0077 | No |
| 63 | Alcam | 20853 | -0.380 | 0.0109 | No |
| 64 | Abca2 | 20921 | -0.385 | 0.0348 | No |
| 65 | Gstm7 | 21085 | -0.395 | 0.0554 | No |
| 66 | Trp53inp1 | 23357 | -0.754 | 0.0113 | No |