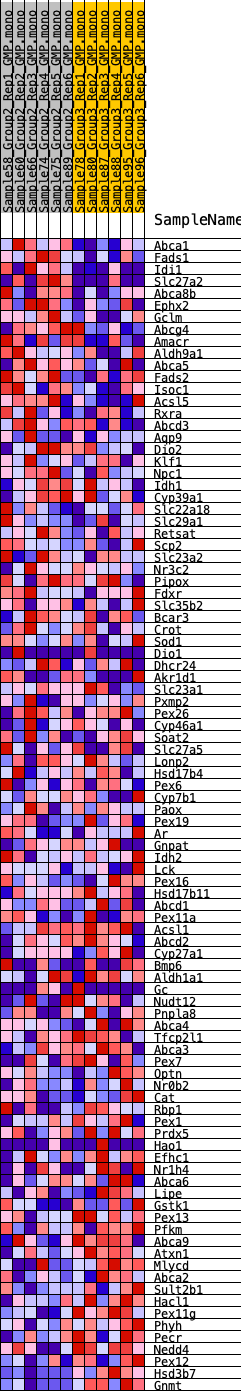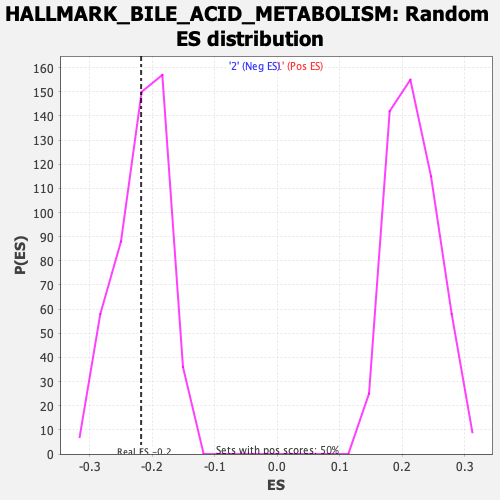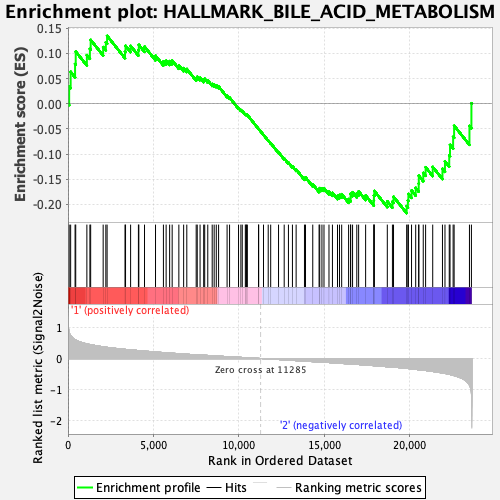
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group2_versus_Group3.GMP.mono_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | GMP.mono_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.21762936 |
| Normalized Enrichment Score (NES) | -1.0042728 |
| Nominal p-value | 0.4233871 |
| FDR q-value | 0.99131155 |
| FWER p-Value | 0.994 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Abca1 | 68 | 0.844 | 0.0344 | No |
| 2 | Fads1 | 155 | 0.740 | 0.0635 | No |
| 3 | Idi1 | 414 | 0.603 | 0.0792 | No |
| 4 | Slc27a2 | 452 | 0.590 | 0.1038 | No |
| 5 | Abca8b | 1104 | 0.469 | 0.0968 | No |
| 6 | Ephx2 | 1281 | 0.450 | 0.1092 | No |
| 7 | Gclm | 1326 | 0.444 | 0.1270 | No |
| 8 | Abcg4 | 2057 | 0.379 | 0.1127 | No |
| 9 | Amacr | 2214 | 0.365 | 0.1222 | No |
| 10 | Aldh9a1 | 2287 | 0.358 | 0.1350 | No |
| 11 | Abca5 | 3335 | 0.295 | 0.1036 | No |
| 12 | Fads2 | 3366 | 0.293 | 0.1153 | No |
| 13 | Isoc1 | 3663 | 0.279 | 0.1150 | No |
| 14 | Acsl5 | 4119 | 0.256 | 0.1070 | No |
| 15 | Rxra | 4144 | 0.254 | 0.1172 | No |
| 16 | Abcd3 | 4476 | 0.237 | 0.1136 | No |
| 17 | Aqp9 | 5120 | 0.209 | 0.0955 | No |
| 18 | Dio2 | 5582 | 0.190 | 0.0843 | No |
| 19 | Klf1 | 5741 | 0.184 | 0.0857 | No |
| 20 | Npc1 | 5946 | 0.175 | 0.0848 | No |
| 21 | Idh1 | 6094 | 0.170 | 0.0861 | No |
| 22 | Cyp39a1 | 6485 | 0.155 | 0.0764 | No |
| 23 | Slc22a18 | 6765 | 0.144 | 0.0709 | No |
| 24 | Slc29a1 | 6950 | 0.138 | 0.0692 | No |
| 25 | Retsat | 7499 | 0.121 | 0.0512 | No |
| 26 | Scp2 | 7561 | 0.118 | 0.0538 | No |
| 27 | Slc23a2 | 7726 | 0.113 | 0.0518 | No |
| 28 | Nr3c2 | 7938 | 0.106 | 0.0475 | No |
| 29 | Pipox | 7997 | 0.104 | 0.0497 | No |
| 30 | Fdxr | 8177 | 0.098 | 0.0464 | No |
| 31 | Slc35b2 | 8436 | 0.089 | 0.0393 | No |
| 32 | Bcar3 | 8551 | 0.085 | 0.0382 | No |
| 33 | Crot | 8675 | 0.081 | 0.0366 | No |
| 34 | Sod1 | 8810 | 0.076 | 0.0343 | No |
| 35 | Dio1 | 9307 | 0.060 | 0.0159 | No |
| 36 | Dhcr24 | 9452 | 0.055 | 0.0122 | No |
| 37 | Akr1d1 | 9977 | 0.039 | -0.0084 | No |
| 38 | Slc23a1 | 10114 | 0.036 | -0.0126 | No |
| 39 | Pxmp2 | 10194 | 0.033 | -0.0145 | No |
| 40 | Pex26 | 10374 | 0.028 | -0.0209 | No |
| 41 | Cyp46a1 | 10412 | 0.027 | -0.0212 | No |
| 42 | Soat2 | 10445 | 0.025 | -0.0215 | No |
| 43 | Slc27a5 | 10493 | 0.024 | -0.0224 | No |
| 44 | Lonp2 | 11146 | 0.004 | -0.0500 | No |
| 45 | Hsd17b4 | 11155 | 0.003 | -0.0502 | No |
| 46 | Pex6 | 11437 | -0.003 | -0.0620 | No |
| 47 | Cyp7b1 | 11713 | -0.013 | -0.0731 | No |
| 48 | Paox | 11858 | -0.017 | -0.0785 | No |
| 49 | Pex19 | 12317 | -0.030 | -0.0966 | No |
| 50 | Ar | 12648 | -0.041 | -0.1088 | No |
| 51 | Gnpat | 12896 | -0.050 | -0.1171 | No |
| 52 | Idh2 | 13130 | -0.058 | -0.1245 | No |
| 53 | Lck | 13346 | -0.066 | -0.1307 | No |
| 54 | Pex16 | 13834 | -0.082 | -0.1478 | No |
| 55 | Hsd17b11 | 13895 | -0.084 | -0.1466 | No |
| 56 | Abcd1 | 14319 | -0.098 | -0.1602 | No |
| 57 | Pex11a | 14689 | -0.108 | -0.1711 | No |
| 58 | Acsl1 | 14727 | -0.110 | -0.1678 | No |
| 59 | Abcd2 | 14856 | -0.115 | -0.1682 | No |
| 60 | Cyp27a1 | 14976 | -0.119 | -0.1680 | No |
| 61 | Bmp6 | 15268 | -0.128 | -0.1747 | No |
| 62 | Aldh1a1 | 15470 | -0.135 | -0.1772 | No |
| 63 | Gc | 15766 | -0.147 | -0.1833 | No |
| 64 | Nudt12 | 15882 | -0.152 | -0.1814 | No |
| 65 | Pnpla8 | 16011 | -0.156 | -0.1800 | No |
| 66 | Abca4 | 16416 | -0.170 | -0.1896 | No |
| 67 | Tfcp2l1 | 16533 | -0.174 | -0.1868 | No |
| 68 | Abca3 | 16534 | -0.174 | -0.1791 | No |
| 69 | Pex7 | 16648 | -0.179 | -0.1760 | No |
| 70 | Optn | 16895 | -0.187 | -0.1782 | No |
| 71 | Nr0b2 | 17003 | -0.192 | -0.1743 | No |
| 72 | Cat | 17412 | -0.209 | -0.1824 | No |
| 73 | Rbp1 | 17892 | -0.229 | -0.1926 | No |
| 74 | Pex1 | 17897 | -0.229 | -0.1827 | No |
| 75 | Prdx5 | 17921 | -0.231 | -0.1734 | No |
| 76 | Hao1 | 18680 | -0.262 | -0.1940 | No |
| 77 | Efhc1 | 18980 | -0.274 | -0.1946 | No |
| 78 | Nr1h4 | 19044 | -0.277 | -0.1850 | No |
| 79 | Abca6 | 19812 | -0.317 | -0.2036 | Yes |
| 80 | Lipe | 19884 | -0.320 | -0.1925 | Yes |
| 81 | Gstk1 | 19914 | -0.321 | -0.1795 | Yes |
| 82 | Pex13 | 20098 | -0.334 | -0.1725 | Yes |
| 83 | Pfkm | 20341 | -0.349 | -0.1674 | Yes |
| 84 | Abca9 | 20517 | -0.360 | -0.1589 | Yes |
| 85 | Atxn1 | 20524 | -0.360 | -0.1432 | Yes |
| 86 | Mlycd | 20783 | -0.375 | -0.1376 | Yes |
| 87 | Abca2 | 20921 | -0.385 | -0.1263 | Yes |
| 88 | Sult2b1 | 21337 | -0.414 | -0.1257 | Yes |
| 89 | Hacl1 | 21907 | -0.464 | -0.1293 | Yes |
| 90 | Pex11g | 22054 | -0.478 | -0.1144 | Yes |
| 91 | Phyh | 22304 | -0.506 | -0.1026 | Yes |
| 92 | Pecr | 22349 | -0.512 | -0.0818 | Yes |
| 93 | Nedd4 | 22531 | -0.542 | -0.0655 | Yes |
| 94 | Pex12 | 22589 | -0.549 | -0.0437 | Yes |
| 95 | Hsd3b7 | 23486 | -0.851 | -0.0441 | Yes |
| 96 | Gnmt | 23601 | -1.129 | 0.0010 | Yes |