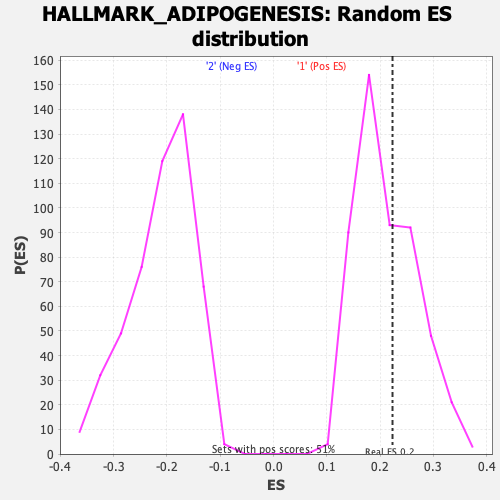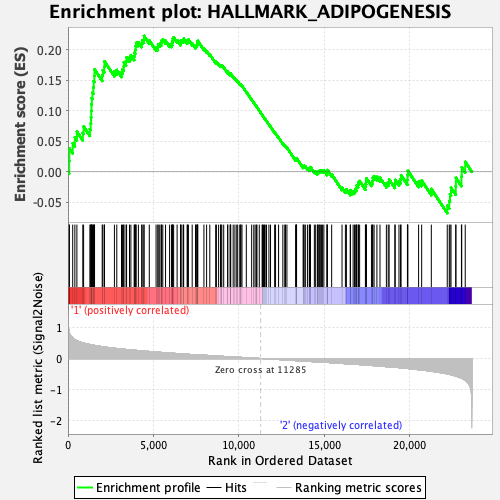
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group2_versus_Group3.GMP.mono_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | GMP.mono_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_ADIPOGENESIS |
| Enrichment Score (ES) | 0.22315033 |
| Normalized Enrichment Score (NES) | 1.0531296 |
| Nominal p-value | 0.39405942 |
| FDR q-value | 0.8794995 |
| FWER p-Value | 0.99 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Abca1 | 68 | 0.844 | 0.0183 | Yes |
| 2 | Plin2 | 81 | 0.821 | 0.0384 | Yes |
| 3 | Lama4 | 275 | 0.662 | 0.0468 | Yes |
| 4 | Sowahc | 401 | 0.605 | 0.0567 | Yes |
| 5 | Gphn | 520 | 0.574 | 0.0661 | Yes |
| 6 | Stom | 866 | 0.504 | 0.0640 | Yes |
| 7 | Esyt1 | 913 | 0.496 | 0.0745 | Yes |
| 8 | Ephx2 | 1281 | 0.450 | 0.0701 | Yes |
| 9 | Gadd45a | 1325 | 0.444 | 0.0795 | Yes |
| 10 | Sparcl1 | 1346 | 0.442 | 0.0897 | Yes |
| 11 | Cidea | 1363 | 0.440 | 0.1001 | Yes |
| 12 | Adipoq | 1374 | 0.440 | 0.1107 | Yes |
| 13 | Aplp2 | 1390 | 0.440 | 0.1211 | Yes |
| 14 | Ppp1r15b | 1432 | 0.435 | 0.1303 | Yes |
| 15 | Itga7 | 1484 | 0.430 | 0.1389 | Yes |
| 16 | Nabp1 | 1503 | 0.427 | 0.1489 | Yes |
| 17 | Idh3a | 1544 | 0.424 | 0.1579 | Yes |
| 18 | Ubqln1 | 1553 | 0.423 | 0.1681 | Yes |
| 19 | Acly | 2001 | 0.383 | 0.1587 | Yes |
| 20 | Dld | 2038 | 0.382 | 0.1667 | Yes |
| 21 | Chuk | 2123 | 0.373 | 0.1725 | Yes |
| 22 | Pfkl | 2136 | 0.371 | 0.1814 | Yes |
| 23 | Me1 | 2715 | 0.332 | 0.1650 | Yes |
| 24 | Tob1 | 2854 | 0.321 | 0.1672 | Yes |
| 25 | Dlat | 3130 | 0.307 | 0.1632 | Yes |
| 26 | Rab34 | 3192 | 0.303 | 0.1682 | Yes |
| 27 | Cdkn2c | 3259 | 0.300 | 0.1729 | Yes |
| 28 | Prdx3 | 3271 | 0.299 | 0.1799 | Yes |
| 29 | Tkt | 3396 | 0.291 | 0.1820 | Yes |
| 30 | Mdh2 | 3427 | 0.290 | 0.1880 | Yes |
| 31 | Cd151 | 3598 | 0.283 | 0.1878 | Yes |
| 32 | Rmdn3 | 3679 | 0.279 | 0.1914 | Yes |
| 33 | Cavin1 | 3870 | 0.268 | 0.1901 | Yes |
| 34 | Mylk | 3893 | 0.267 | 0.1958 | Yes |
| 35 | Aco2 | 3939 | 0.265 | 0.2006 | Yes |
| 36 | G3bp2 | 3962 | 0.264 | 0.2062 | Yes |
| 37 | C3 | 3986 | 0.263 | 0.2119 | Yes |
| 38 | Grpel1 | 4118 | 0.256 | 0.2127 | Yes |
| 39 | Uqcr11 | 4310 | 0.245 | 0.2107 | Yes |
| 40 | Ddt | 4337 | 0.244 | 0.2157 | Yes |
| 41 | Ifngr1 | 4441 | 0.239 | 0.2173 | Yes |
| 42 | Ywhag | 4446 | 0.238 | 0.2232 | Yes |
| 43 | Aldoa | 4756 | 0.227 | 0.2157 | No |
| 44 | Pemt | 5153 | 0.208 | 0.2040 | No |
| 45 | Sqor | 5259 | 0.203 | 0.2046 | No |
| 46 | Mrpl15 | 5271 | 0.202 | 0.2092 | No |
| 47 | Tst | 5366 | 0.199 | 0.2102 | No |
| 48 | Ptcd3 | 5448 | 0.196 | 0.2117 | No |
| 49 | Mtch2 | 5474 | 0.194 | 0.2155 | No |
| 50 | Lifr | 5545 | 0.191 | 0.2173 | No |
| 51 | Atp1b3 | 5706 | 0.185 | 0.2151 | No |
| 52 | Ak2 | 5944 | 0.175 | 0.2094 | No |
| 53 | Immt | 6058 | 0.171 | 0.2089 | No |
| 54 | Idh1 | 6094 | 0.170 | 0.2117 | No |
| 55 | Sorbs1 | 6115 | 0.169 | 0.2151 | No |
| 56 | Mccc1 | 6131 | 0.168 | 0.2186 | No |
| 57 | Cs | 6180 | 0.167 | 0.2208 | No |
| 58 | Cmpk1 | 6383 | 0.159 | 0.2162 | No |
| 59 | Itsn1 | 6583 | 0.152 | 0.2115 | No |
| 60 | Uqcrq | 6585 | 0.152 | 0.2152 | No |
| 61 | Fzd4 | 6659 | 0.149 | 0.2159 | No |
| 62 | Reep5 | 6748 | 0.145 | 0.2157 | No |
| 63 | Acadl | 6767 | 0.144 | 0.2186 | No |
| 64 | Ndufs3 | 6969 | 0.137 | 0.2135 | No |
| 65 | Ghitm | 6993 | 0.136 | 0.2159 | No |
| 66 | Apoe | 7045 | 0.134 | 0.2171 | No |
| 67 | Gpam | 7265 | 0.125 | 0.2109 | No |
| 68 | Suclg1 | 7459 | 0.122 | 0.2058 | No |
| 69 | Retsat | 7499 | 0.121 | 0.2071 | No |
| 70 | Mtarc2 | 7543 | 0.119 | 0.2083 | No |
| 71 | Scp2 | 7561 | 0.118 | 0.2105 | No |
| 72 | Lpcat3 | 7576 | 0.117 | 0.2129 | No |
| 73 | Preb | 7595 | 0.117 | 0.2150 | No |
| 74 | Ptger3 | 7952 | 0.105 | 0.2025 | No |
| 75 | Elovl6 | 8109 | 0.100 | 0.1983 | No |
| 76 | Rtn3 | 8296 | 0.093 | 0.1927 | No |
| 77 | Riok3 | 8640 | 0.082 | 0.1802 | No |
| 78 | Cd302 | 8682 | 0.081 | 0.1804 | No |
| 79 | Sod1 | 8810 | 0.076 | 0.1769 | No |
| 80 | Pfkfb3 | 8921 | 0.073 | 0.1741 | No |
| 81 | Gpat4 | 8954 | 0.072 | 0.1745 | No |
| 82 | Esrra | 8998 | 0.070 | 0.1744 | No |
| 83 | Uqcrc1 | 9109 | 0.067 | 0.1714 | No |
| 84 | Scarb1 | 9343 | 0.059 | 0.1630 | No |
| 85 | Etfb | 9344 | 0.059 | 0.1644 | No |
| 86 | Cox7b | 9484 | 0.054 | 0.1599 | No |
| 87 | Samm50 | 9502 | 0.054 | 0.1605 | No |
| 88 | Angptl4 | 9520 | 0.053 | 0.1611 | No |
| 89 | Atl2 | 9663 | 0.048 | 0.1563 | No |
| 90 | Ppm1b | 9765 | 0.045 | 0.1531 | No |
| 91 | Ech1 | 9872 | 0.042 | 0.1496 | No |
| 92 | Gpx4 | 9929 | 0.041 | 0.1483 | No |
| 93 | Sdhb | 10048 | 0.038 | 0.1442 | No |
| 94 | Ccng2 | 10102 | 0.036 | 0.1428 | No |
| 95 | Aifm1 | 10112 | 0.036 | 0.1433 | No |
| 96 | Dhrs7b | 10178 | 0.034 | 0.1414 | No |
| 97 | Jagn1 | 10432 | 0.026 | 0.1312 | No |
| 98 | Cox6a1 | 10744 | 0.016 | 0.1184 | No |
| 99 | Ubc | 10869 | 0.013 | 0.1134 | No |
| 100 | Bcl2l13 | 10949 | 0.010 | 0.1103 | No |
| 101 | Tank | 11034 | 0.007 | 0.1069 | No |
| 102 | Slc19a1 | 11052 | 0.007 | 0.1063 | No |
| 103 | Ucp2 | 11190 | 0.002 | 0.1005 | No |
| 104 | Ndufab1 | 11357 | -0.000 | 0.0935 | No |
| 105 | Nmt1 | 11377 | -0.001 | 0.0927 | No |
| 106 | Slc5a6 | 11439 | -0.003 | 0.0901 | No |
| 107 | Dram2 | 11495 | -0.005 | 0.0879 | No |
| 108 | Abcb8 | 11534 | -0.006 | 0.0864 | No |
| 109 | Col4a1 | 11614 | -0.009 | 0.0833 | No |
| 110 | Qdpr | 11754 | -0.014 | 0.0777 | No |
| 111 | Slc25a10 | 11848 | -0.017 | 0.0742 | No |
| 112 | Cpt2 | 12090 | -0.024 | 0.0645 | No |
| 113 | Pex14 | 12142 | -0.025 | 0.0629 | No |
| 114 | Rnf11 | 12319 | -0.030 | 0.0562 | No |
| 115 | Mrap | 12569 | -0.038 | 0.0465 | No |
| 116 | Arl4a | 12681 | -0.042 | 0.0428 | No |
| 117 | Dgat1 | 12723 | -0.043 | 0.0422 | No |
| 118 | Rreb1 | 12808 | -0.046 | 0.0398 | No |
| 119 | Itih5 | 13316 | -0.065 | 0.0198 | No |
| 120 | Map4k3 | 13350 | -0.066 | 0.0200 | No |
| 121 | Acadm | 13367 | -0.066 | 0.0210 | No |
| 122 | Por | 13372 | -0.066 | 0.0225 | No |
| 123 | Uck1 | 13755 | -0.080 | 0.0082 | No |
| 124 | Gpd2 | 13791 | -0.081 | 0.0087 | No |
| 125 | Uqcr10 | 13810 | -0.081 | 0.0100 | No |
| 126 | Cmbl | 13885 | -0.084 | 0.0089 | No |
| 127 | Cyc1 | 14007 | -0.088 | 0.0060 | No |
| 128 | Gpx3 | 14114 | -0.091 | 0.0037 | No |
| 129 | Mgll | 14128 | -0.092 | 0.0055 | No |
| 130 | Lpl | 14165 | -0.093 | 0.0063 | No |
| 131 | Slc1a5 | 14185 | -0.094 | 0.0078 | No |
| 132 | Dhcr7 | 14413 | -0.101 | 0.0007 | No |
| 133 | Taldo1 | 14489 | -0.102 | 0.0000 | No |
| 134 | Pdcd4 | 14592 | -0.105 | -0.0017 | No |
| 135 | Acox1 | 14612 | -0.106 | 0.0002 | No |
| 136 | Agpat3 | 14672 | -0.108 | 0.0004 | No |
| 137 | Ndufa5 | 14703 | -0.109 | 0.0018 | No |
| 138 | Crat | 14761 | -0.112 | 0.0022 | No |
| 139 | Dhrs7 | 14826 | -0.114 | 0.0023 | No |
| 140 | Dnajb9 | 14894 | -0.116 | 0.0024 | No |
| 141 | Baz2a | 14969 | -0.119 | 0.0022 | No |
| 142 | Dnajc15 | 15153 | -0.125 | -0.0024 | No |
| 143 | Nkiras1 | 15156 | -0.125 | 0.0006 | No |
| 144 | Ndufb7 | 15176 | -0.126 | 0.0030 | No |
| 145 | Slc25a1 | 15425 | -0.134 | -0.0042 | No |
| 146 | Phldb1 | 16031 | -0.157 | -0.0261 | No |
| 147 | Reep6 | 16229 | -0.164 | -0.0304 | No |
| 148 | Adipor2 | 16287 | -0.166 | -0.0287 | No |
| 149 | Bcl6 | 16514 | -0.173 | -0.0340 | No |
| 150 | Cyp4b1 | 16522 | -0.173 | -0.0299 | No |
| 151 | Stat5a | 16681 | -0.180 | -0.0321 | No |
| 152 | Chchd10 | 16764 | -0.182 | -0.0311 | No |
| 153 | Cd36 | 16808 | -0.183 | -0.0283 | No |
| 154 | Pparg | 16877 | -0.186 | -0.0265 | No |
| 155 | Pim3 | 16888 | -0.186 | -0.0223 | No |
| 156 | Cavin2 | 16972 | -0.191 | -0.0210 | No |
| 157 | Acaa2 | 16997 | -0.192 | -0.0172 | No |
| 158 | Mgst3 | 17061 | -0.194 | -0.0150 | No |
| 159 | Gbe1 | 17399 | -0.208 | -0.0242 | No |
| 160 | Cat | 17412 | -0.209 | -0.0194 | No |
| 161 | Echs1 | 17440 | -0.210 | -0.0153 | No |
| 162 | Hadh | 17459 | -0.211 | -0.0108 | No |
| 163 | Slc27a1 | 17753 | -0.222 | -0.0177 | No |
| 164 | Coq3 | 17818 | -0.225 | -0.0148 | No |
| 165 | Hibch | 17831 | -0.226 | -0.0096 | No |
| 166 | Sdhc | 17906 | -0.230 | -0.0070 | No |
| 167 | Sspn | 18072 | -0.237 | -0.0081 | No |
| 168 | Miga2 | 18252 | -0.244 | -0.0096 | No |
| 169 | Col15a1 | 18626 | -0.261 | -0.0189 | No |
| 170 | Pgm1 | 18732 | -0.264 | -0.0168 | No |
| 171 | Aldh2 | 18787 | -0.267 | -0.0124 | No |
| 172 | Idh3g | 19122 | -0.282 | -0.0196 | No |
| 173 | Coq5 | 19141 | -0.283 | -0.0132 | No |
| 174 | Cox8a | 19363 | -0.292 | -0.0153 | No |
| 175 | Araf | 19455 | -0.297 | -0.0117 | No |
| 176 | Adcy6 | 19484 | -0.299 | -0.0054 | No |
| 177 | Enpp2 | 19858 | -0.319 | -0.0133 | No |
| 178 | Fah | 19863 | -0.319 | -0.0055 | No |
| 179 | Lipe | 19884 | -0.320 | 0.0017 | No |
| 180 | Coq9 | 20511 | -0.360 | -0.0160 | No |
| 181 | Acads | 20690 | -0.370 | -0.0143 | No |
| 182 | Decr1 | 21255 | -0.407 | -0.0281 | No |
| 183 | Dbt | 22187 | -0.493 | -0.0554 | No |
| 184 | Phyh | 22304 | -0.506 | -0.0477 | No |
| 185 | Omd | 22348 | -0.512 | -0.0366 | No |
| 186 | Vegfb | 22407 | -0.522 | -0.0260 | No |
| 187 | Elmod3 | 22685 | -0.563 | -0.0237 | No |
| 188 | Bckdha | 22693 | -0.563 | -0.0098 | No |
| 189 | Angpt1 | 23017 | -0.629 | -0.0078 | No |
| 190 | Ltc4s | 23034 | -0.635 | 0.0075 | No |
| 191 | Sult1a1 | 23239 | -0.702 | 0.0164 | No |