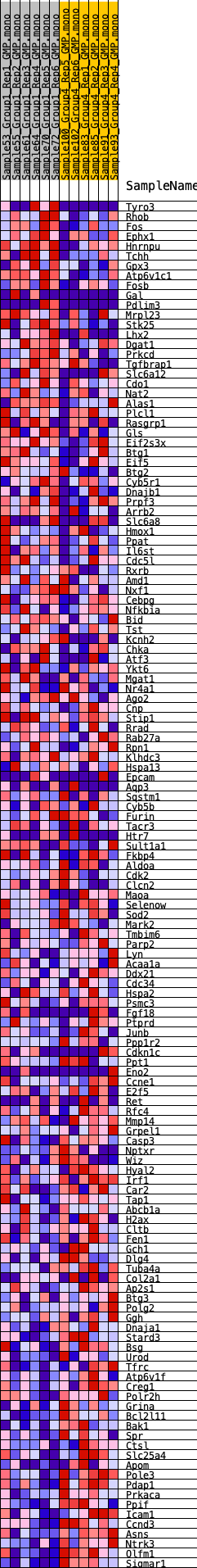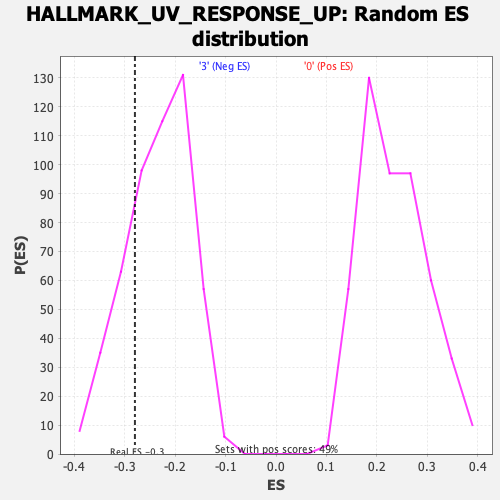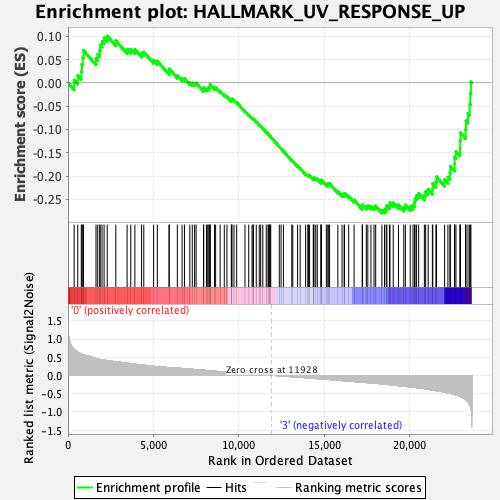
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group4.GMP.mono_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_UV_RESPONSE_UP |
| Enrichment Score (ES) | -0.2794553 |
| Normalized Enrichment Score (NES) | -1.1979606 |
| Nominal p-value | 0.24366471 |
| FDR q-value | 0.49653292 |
| FWER p-Value | 0.924 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tyro3 | 360 | 0.726 | 0.0061 | No |
| 2 | Rhob | 570 | 0.638 | 0.0160 | No |
| 3 | Fos | 777 | 0.588 | 0.0246 | No |
| 4 | Ephx1 | 813 | 0.580 | 0.0402 | No |
| 5 | Hnrnpu | 866 | 0.569 | 0.0548 | No |
| 6 | Tchh | 905 | 0.564 | 0.0698 | No |
| 7 | Gpx3 | 1638 | 0.464 | 0.0523 | No |
| 8 | Atp6v1c1 | 1739 | 0.454 | 0.0614 | No |
| 9 | Fosb | 1857 | 0.439 | 0.0694 | No |
| 10 | Gal | 1882 | 0.436 | 0.0813 | No |
| 11 | Pdlim3 | 1993 | 0.427 | 0.0892 | No |
| 12 | Mrpl23 | 2112 | 0.419 | 0.0965 | No |
| 13 | Stk25 | 2294 | 0.406 | 0.1008 | No |
| 14 | Lhx2 | 2798 | 0.373 | 0.0904 | No |
| 15 | Dgat1 | 3458 | 0.336 | 0.0722 | No |
| 16 | Prkcd | 3674 | 0.320 | 0.0725 | No |
| 17 | Tgfbrap1 | 3912 | 0.308 | 0.0715 | No |
| 18 | Slc6a12 | 4304 | 0.287 | 0.0633 | No |
| 19 | Cdo1 | 4434 | 0.280 | 0.0661 | No |
| 20 | Nat2 | 5009 | 0.250 | 0.0490 | No |
| 21 | Alas1 | 5226 | 0.239 | 0.0469 | No |
| 22 | Plcl1 | 5916 | 0.216 | 0.0239 | No |
| 23 | Rasgrp1 | 5917 | 0.216 | 0.0303 | No |
| 24 | Gls | 6396 | 0.198 | 0.0158 | No |
| 25 | Eif2s3x | 6675 | 0.191 | 0.0096 | No |
| 26 | Btg1 | 6813 | 0.185 | 0.0092 | No |
| 27 | Eif5 | 7127 | 0.173 | 0.0010 | No |
| 28 | Btg2 | 7271 | 0.168 | -0.0001 | No |
| 29 | Cyb5r1 | 7406 | 0.162 | -0.0010 | No |
| 30 | Dnajb1 | 7494 | 0.159 | -0.0000 | No |
| 31 | Prpf3 | 7931 | 0.143 | -0.0144 | No |
| 32 | Arrb2 | 7938 | 0.143 | -0.0104 | No |
| 33 | Slc6a8 | 8100 | 0.135 | -0.0133 | No |
| 34 | Hmox1 | 8175 | 0.132 | -0.0125 | No |
| 35 | Ppat | 8247 | 0.130 | -0.0117 | No |
| 36 | Il6st | 8258 | 0.129 | -0.0084 | No |
| 37 | Cdc5l | 8314 | 0.127 | -0.0069 | No |
| 38 | Rxrb | 8320 | 0.127 | -0.0034 | No |
| 39 | Amd1 | 8574 | 0.117 | -0.0107 | No |
| 40 | Nxf1 | 8637 | 0.114 | -0.0100 | No |
| 41 | Cebpg | 8900 | 0.105 | -0.0181 | No |
| 42 | Nfkbia | 9143 | 0.096 | -0.0255 | No |
| 43 | Bid | 9298 | 0.091 | -0.0294 | No |
| 44 | Tst | 9554 | 0.082 | -0.0378 | No |
| 45 | Kcnh2 | 9574 | 0.082 | -0.0362 | No |
| 46 | Chka | 9591 | 0.081 | -0.0345 | No |
| 47 | Atf3 | 9711 | 0.077 | -0.0373 | No |
| 48 | Ykt6 | 9868 | 0.072 | -0.0418 | No |
| 49 | Mgat1 | 10353 | 0.054 | -0.0609 | No |
| 50 | Nr4a1 | 10571 | 0.045 | -0.0688 | No |
| 51 | Ago2 | 10767 | 0.038 | -0.0759 | No |
| 52 | Cnp | 10828 | 0.036 | -0.0775 | No |
| 53 | Stip1 | 10853 | 0.035 | -0.0775 | No |
| 54 | Rrad | 11014 | 0.029 | -0.0834 | No |
| 55 | Rab27a | 11190 | 0.024 | -0.0901 | No |
| 56 | Rpn1 | 11273 | 0.022 | -0.0930 | No |
| 57 | Klhdc3 | 11398 | 0.018 | -0.0977 | No |
| 58 | Hspa13 | 11597 | 0.011 | -0.1058 | No |
| 59 | Epcam | 11700 | 0.008 | -0.1099 | No |
| 60 | Aqp3 | 11743 | 0.006 | -0.1116 | No |
| 61 | Sqstm1 | 11763 | 0.005 | -0.1122 | No |
| 62 | Cyb5b | 11805 | 0.004 | -0.1138 | No |
| 63 | Furin | 11818 | 0.004 | -0.1142 | No |
| 64 | Tacr3 | 11861 | 0.002 | -0.1160 | No |
| 65 | Htr7 | 12360 | -0.006 | -0.1370 | No |
| 66 | Sult1a1 | 12465 | -0.010 | -0.1411 | No |
| 67 | Fkbp4 | 12603 | -0.014 | -0.1465 | No |
| 68 | Aldoa | 13091 | -0.031 | -0.1664 | No |
| 69 | Cdk2 | 13151 | -0.033 | -0.1679 | No |
| 70 | Clcn2 | 13432 | -0.044 | -0.1785 | No |
| 71 | Maoa | 13582 | -0.049 | -0.1834 | No |
| 72 | Selenow | 13902 | -0.059 | -0.1952 | No |
| 73 | Sod2 | 14029 | -0.064 | -0.1987 | No |
| 74 | Mark2 | 14077 | -0.065 | -0.1988 | No |
| 75 | Tmbim6 | 14132 | -0.068 | -0.1991 | No |
| 76 | Parp2 | 14349 | -0.076 | -0.2060 | No |
| 77 | Lyn | 14365 | -0.077 | -0.2044 | No |
| 78 | Acaa1a | 14390 | -0.078 | -0.2031 | No |
| 79 | Ddx21 | 14493 | -0.081 | -0.2051 | No |
| 80 | Cdc34 | 14585 | -0.085 | -0.2065 | No |
| 81 | Hspa2 | 14798 | -0.091 | -0.2128 | No |
| 82 | Psmc3 | 14812 | -0.092 | -0.2106 | No |
| 83 | Fgf18 | 14817 | -0.092 | -0.2081 | No |
| 84 | Ptprd | 15115 | -0.102 | -0.2177 | No |
| 85 | Junb | 15208 | -0.107 | -0.2185 | No |
| 86 | Ppp1r2 | 15222 | -0.107 | -0.2159 | No |
| 87 | Cdkn1c | 15304 | -0.110 | -0.2161 | No |
| 88 | Ppt1 | 15788 | -0.128 | -0.2329 | No |
| 89 | Eno2 | 16033 | -0.138 | -0.2392 | No |
| 90 | Ccne1 | 16154 | -0.142 | -0.2401 | No |
| 91 | E2f5 | 16177 | -0.143 | -0.2368 | No |
| 92 | Ret | 16421 | -0.153 | -0.2427 | No |
| 93 | Rfc4 | 16736 | -0.164 | -0.2512 | No |
| 94 | Mmp14 | 17211 | -0.181 | -0.2660 | No |
| 95 | Grpel1 | 17229 | -0.182 | -0.2614 | No |
| 96 | Casp3 | 17452 | -0.191 | -0.2652 | No |
| 97 | Nptxr | 17541 | -0.196 | -0.2632 | No |
| 98 | Wiz | 17724 | -0.203 | -0.2649 | No |
| 99 | Hyal2 | 17902 | -0.211 | -0.2662 | No |
| 100 | Irf1 | 18000 | -0.215 | -0.2640 | No |
| 101 | Car2 | 18364 | -0.228 | -0.2727 | Yes |
| 102 | Tap1 | 18516 | -0.234 | -0.2722 | Yes |
| 103 | Abcb1a | 18615 | -0.239 | -0.2693 | Yes |
| 104 | H2ax | 18654 | -0.241 | -0.2639 | Yes |
| 105 | Cltb | 18805 | -0.249 | -0.2629 | Yes |
| 106 | Fen1 | 18837 | -0.252 | -0.2568 | Yes |
| 107 | Gch1 | 19027 | -0.262 | -0.2571 | Yes |
| 108 | Dlg4 | 19333 | -0.276 | -0.2619 | Yes |
| 109 | Tuba4a | 19643 | -0.291 | -0.2665 | Yes |
| 110 | Col2a1 | 19737 | -0.296 | -0.2617 | Yes |
| 111 | Ap2s1 | 20022 | -0.312 | -0.2646 | Yes |
| 112 | Btg3 | 20184 | -0.319 | -0.2621 | Yes |
| 113 | Polg2 | 20273 | -0.323 | -0.2563 | Yes |
| 114 | Ggh | 20301 | -0.325 | -0.2478 | Yes |
| 115 | Dnaja1 | 20384 | -0.330 | -0.2416 | Yes |
| 116 | Stard3 | 20514 | -0.338 | -0.2371 | Yes |
| 117 | Bsg | 20848 | -0.359 | -0.2407 | Yes |
| 118 | Urod | 20924 | -0.365 | -0.2331 | Yes |
| 119 | Tfrc | 21073 | -0.375 | -0.2284 | Yes |
| 120 | Atp6v1f | 21317 | -0.393 | -0.2271 | Yes |
| 121 | Creg1 | 21341 | -0.396 | -0.2164 | Yes |
| 122 | Polr2h | 21525 | -0.412 | -0.2120 | Yes |
| 123 | Grina | 21560 | -0.415 | -0.2012 | Yes |
| 124 | Bcl2l11 | 22033 | -0.455 | -0.2079 | Yes |
| 125 | Bak1 | 22226 | -0.475 | -0.2021 | Yes |
| 126 | Spr | 22345 | -0.488 | -0.1927 | Yes |
| 127 | Ctsl | 22369 | -0.490 | -0.1792 | Yes |
| 128 | Slc25a4 | 22615 | -0.520 | -0.1743 | Yes |
| 129 | Apom | 22620 | -0.520 | -0.1592 | Yes |
| 130 | Pole3 | 22696 | -0.531 | -0.1467 | Yes |
| 131 | Pdap1 | 22933 | -0.573 | -0.1398 | Yes |
| 132 | Prkaca | 22942 | -0.575 | -0.1232 | Yes |
| 133 | Ppif | 22960 | -0.578 | -0.1069 | Yes |
| 134 | Icam1 | 23259 | -0.657 | -0.1002 | Yes |
| 135 | Ccnd3 | 23283 | -0.667 | -0.0815 | Yes |
| 136 | Asns | 23402 | -0.716 | -0.0654 | Yes |
| 137 | Ntrk3 | 23500 | -0.796 | -0.0460 | Yes |
| 138 | Olfm1 | 23539 | -0.839 | -0.0229 | Yes |
| 139 | Sigmar1 | 23571 | -0.897 | 0.0023 | Yes |