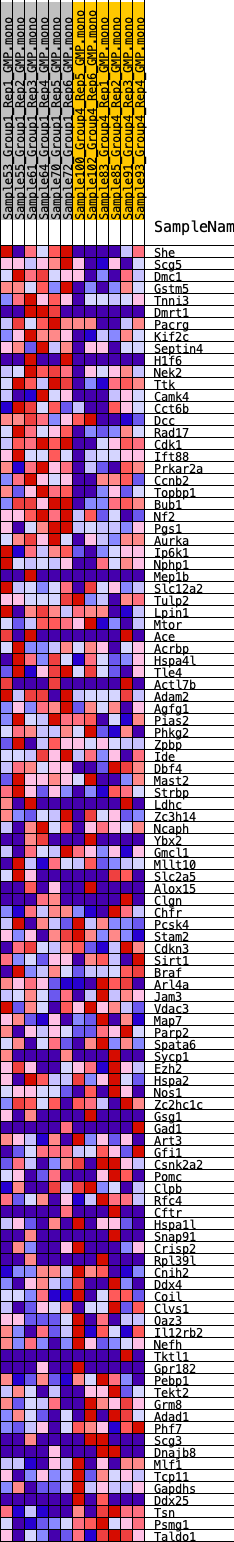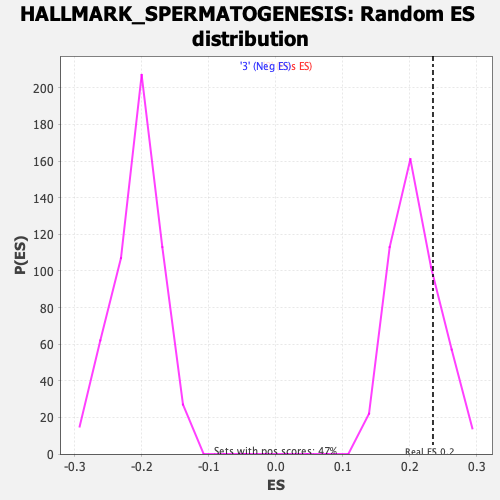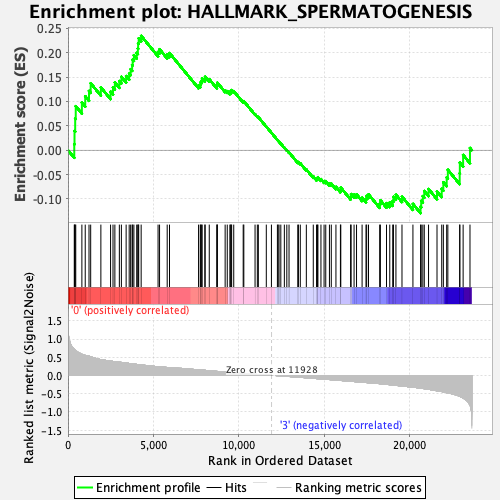
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group4.GMP.mono_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | 0.23454449 |
| Normalized Enrichment Score (NES) | 1.1322798 |
| Nominal p-value | 0.21108742 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.953 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | She | 367 | 0.721 | 0.0124 | Yes |
| 2 | Scg5 | 381 | 0.715 | 0.0396 | Yes |
| 3 | Dmc1 | 421 | 0.698 | 0.0650 | Yes |
| 4 | Gstm5 | 453 | 0.684 | 0.0903 | Yes |
| 5 | Tnni3 | 812 | 0.580 | 0.0976 | Yes |
| 6 | Dmrt1 | 1008 | 0.549 | 0.1106 | Yes |
| 7 | Pacrg | 1229 | 0.525 | 0.1216 | Yes |
| 8 | Kif2c | 1327 | 0.509 | 0.1373 | Yes |
| 9 | Septin4 | 1923 | 0.434 | 0.1288 | Yes |
| 10 | H1f6 | 2490 | 0.391 | 0.1200 | Yes |
| 11 | Nek2 | 2633 | 0.383 | 0.1288 | Yes |
| 12 | Ttk | 2742 | 0.379 | 0.1389 | Yes |
| 13 | Camk4 | 3004 | 0.363 | 0.1419 | Yes |
| 14 | Cct6b | 3123 | 0.356 | 0.1507 | Yes |
| 15 | Dcc | 3401 | 0.340 | 0.1521 | Yes |
| 16 | Rad17 | 3579 | 0.328 | 0.1573 | Yes |
| 17 | Cdk1 | 3665 | 0.321 | 0.1662 | Yes |
| 18 | Ift88 | 3752 | 0.318 | 0.1749 | Yes |
| 19 | Prkar2a | 3783 | 0.317 | 0.1859 | Yes |
| 20 | Ccnb2 | 3855 | 0.311 | 0.1949 | Yes |
| 21 | Topbp1 | 4020 | 0.302 | 0.1997 | Yes |
| 22 | Bub1 | 4087 | 0.298 | 0.2085 | Yes |
| 23 | Nf2 | 4103 | 0.297 | 0.2194 | Yes |
| 24 | Pgs1 | 4136 | 0.295 | 0.2294 | Yes |
| 25 | Aurka | 4280 | 0.288 | 0.2345 | Yes |
| 26 | Ip6k1 | 5269 | 0.237 | 0.2017 | No |
| 27 | Nphp1 | 5356 | 0.233 | 0.2071 | No |
| 28 | Mep1b | 5803 | 0.218 | 0.1966 | No |
| 29 | Slc12a2 | 5938 | 0.215 | 0.1992 | No |
| 30 | Tulp2 | 7638 | 0.154 | 0.1330 | No |
| 31 | Lpin1 | 7737 | 0.151 | 0.1347 | No |
| 32 | Mtor | 7761 | 0.150 | 0.1395 | No |
| 33 | Ace | 7818 | 0.147 | 0.1428 | No |
| 34 | Acrbp | 7846 | 0.147 | 0.1474 | No |
| 35 | Hspa4l | 8008 | 0.140 | 0.1460 | No |
| 36 | Tle4 | 8021 | 0.139 | 0.1509 | No |
| 37 | Actl7b | 8264 | 0.129 | 0.1456 | No |
| 38 | Adam2 | 8710 | 0.111 | 0.1310 | No |
| 39 | Agfg1 | 8723 | 0.110 | 0.1347 | No |
| 40 | Pias2 | 8735 | 0.110 | 0.1385 | No |
| 41 | Phkg2 | 9193 | 0.095 | 0.1228 | No |
| 42 | Zpbp | 9317 | 0.091 | 0.1211 | No |
| 43 | Ide | 9470 | 0.086 | 0.1179 | No |
| 44 | Dbf4 | 9525 | 0.083 | 0.1189 | No |
| 45 | Mast2 | 9529 | 0.083 | 0.1220 | No |
| 46 | Strbp | 9573 | 0.082 | 0.1233 | No |
| 47 | Ldhc | 9696 | 0.078 | 0.1211 | No |
| 48 | Zc3h14 | 10249 | 0.057 | 0.0999 | No |
| 49 | Ncaph | 10286 | 0.056 | 0.1005 | No |
| 50 | Ybx2 | 10943 | 0.031 | 0.0738 | No |
| 51 | Gmcl1 | 11077 | 0.027 | 0.0692 | No |
| 52 | Mllt10 | 11136 | 0.025 | 0.0677 | No |
| 53 | Slc2a5 | 11601 | 0.011 | 0.0484 | No |
| 54 | Alox15 | 11901 | 0.001 | 0.0358 | No |
| 55 | Clgn | 12250 | -0.002 | 0.0211 | No |
| 56 | Chfr | 12290 | -0.004 | 0.0195 | No |
| 57 | Pcsk4 | 12345 | -0.006 | 0.0175 | No |
| 58 | Stam2 | 12450 | -0.009 | 0.0134 | No |
| 59 | Cdkn3 | 12657 | -0.016 | 0.0053 | No |
| 60 | Sirt1 | 12801 | -0.021 | 0.0000 | No |
| 61 | Braf | 12931 | -0.026 | -0.0045 | No |
| 62 | Arl4a | 13443 | -0.044 | -0.0245 | No |
| 63 | Jam3 | 13481 | -0.046 | -0.0243 | No |
| 64 | Vdac3 | 13590 | -0.049 | -0.0269 | No |
| 65 | Map7 | 13943 | -0.061 | -0.0396 | No |
| 66 | Parp2 | 14349 | -0.076 | -0.0538 | No |
| 67 | Spata6 | 14541 | -0.083 | -0.0587 | No |
| 68 | Sycp1 | 14593 | -0.085 | -0.0576 | No |
| 69 | Ezh2 | 14621 | -0.085 | -0.0554 | No |
| 70 | Hspa2 | 14798 | -0.091 | -0.0594 | No |
| 71 | Nos1 | 14981 | -0.098 | -0.0633 | No |
| 72 | Zc2hc1c | 15079 | -0.101 | -0.0635 | No |
| 73 | Gsg1 | 15295 | -0.110 | -0.0683 | No |
| 74 | Gad1 | 15403 | -0.114 | -0.0685 | No |
| 75 | Art3 | 15675 | -0.123 | -0.0752 | No |
| 76 | Gfi1 | 15937 | -0.134 | -0.0811 | No |
| 77 | Csnk2a2 | 15959 | -0.135 | -0.0768 | No |
| 78 | Pomc | 16534 | -0.158 | -0.0951 | No |
| 79 | Clpb | 16561 | -0.158 | -0.0900 | No |
| 80 | Rfc4 | 16736 | -0.164 | -0.0910 | No |
| 81 | Cftr | 16880 | -0.170 | -0.0905 | No |
| 82 | Hspa1l | 17205 | -0.181 | -0.0973 | No |
| 83 | Snap91 | 17434 | -0.191 | -0.0996 | No |
| 84 | Crisp2 | 17473 | -0.193 | -0.0937 | No |
| 85 | Rpl39l | 17580 | -0.197 | -0.0906 | No |
| 86 | Cnih2 | 18234 | -0.222 | -0.1097 | No |
| 87 | Ddx4 | 18275 | -0.224 | -0.1027 | No |
| 88 | Coil | 18634 | -0.240 | -0.1086 | No |
| 89 | Clvs1 | 18824 | -0.251 | -0.1069 | No |
| 90 | Oaz3 | 18998 | -0.260 | -0.1042 | No |
| 91 | Il12rb2 | 19040 | -0.263 | -0.0957 | No |
| 92 | Nefh | 19185 | -0.269 | -0.0914 | No |
| 93 | Tktl1 | 19539 | -0.285 | -0.0953 | No |
| 94 | Gpr182 | 20179 | -0.318 | -0.1101 | No |
| 95 | Pebp1 | 20626 | -0.345 | -0.1157 | No |
| 96 | Tekt2 | 20663 | -0.347 | -0.1038 | No |
| 97 | Grm8 | 20759 | -0.353 | -0.0941 | No |
| 98 | Adad1 | 20843 | -0.359 | -0.0837 | No |
| 99 | Phf7 | 21093 | -0.377 | -0.0796 | No |
| 100 | Scg3 | 21588 | -0.418 | -0.0844 | No |
| 101 | Dnajb8 | 21864 | -0.438 | -0.0791 | No |
| 102 | Mlf1 | 21959 | -0.448 | -0.0657 | No |
| 103 | Tcp11 | 22154 | -0.468 | -0.0558 | No |
| 104 | Gapdhs | 22222 | -0.475 | -0.0402 | No |
| 105 | Ddx25 | 22915 | -0.571 | -0.0475 | No |
| 106 | Tsn | 22924 | -0.572 | -0.0256 | No |
| 107 | Psmg1 | 23116 | -0.617 | -0.0098 | No |
| 108 | Taldo1 | 23517 | -0.806 | 0.0045 | No |