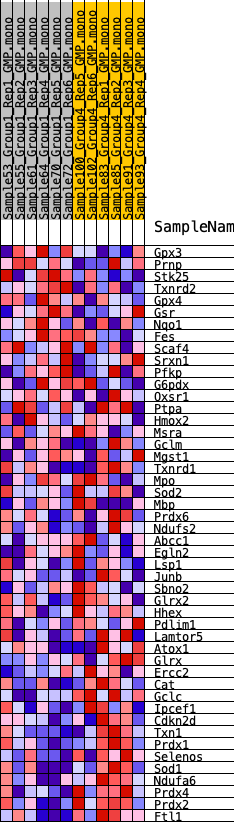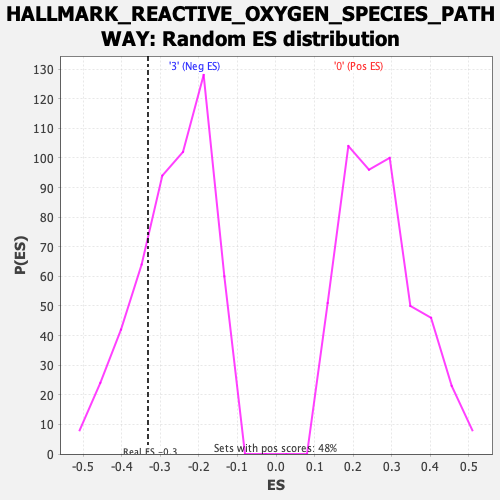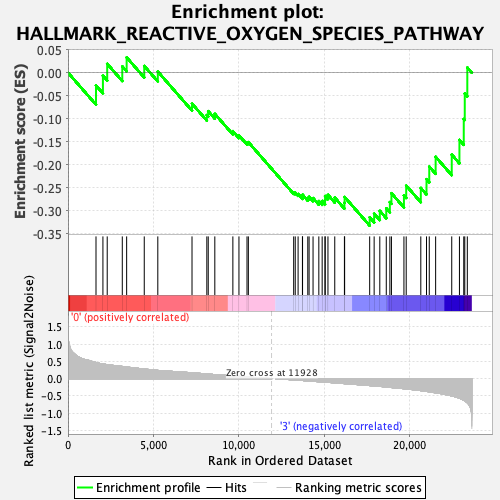
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group4.GMP.mono_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_REACTIVE_OXYGEN_SPECIES_PATHWAY |
| Enrichment Score (ES) | -0.33216488 |
| Normalized Enrichment Score (NES) | -1.2503861 |
| Nominal p-value | 0.25095785 |
| FDR q-value | 0.4134616 |
| FWER p-Value | 0.862 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gpx3 | 1638 | 0.464 | -0.0277 | No |
| 2 | Prnp | 2043 | 0.424 | -0.0067 | No |
| 3 | Stk25 | 2294 | 0.406 | 0.0192 | No |
| 4 | Txnrd2 | 3176 | 0.352 | 0.0135 | No |
| 5 | Gpx4 | 3431 | 0.338 | 0.0331 | No |
| 6 | Gsr | 4461 | 0.278 | 0.0145 | No |
| 7 | Nqo1 | 5254 | 0.238 | 0.0023 | No |
| 8 | Fes | 7255 | 0.168 | -0.0673 | No |
| 9 | Scaf4 | 8122 | 0.134 | -0.0920 | No |
| 10 | Srxn1 | 8204 | 0.131 | -0.0836 | No |
| 11 | Pfkp | 8589 | 0.116 | -0.0895 | No |
| 12 | G6pdx | 9645 | 0.080 | -0.1271 | No |
| 13 | Oxsr1 | 10001 | 0.066 | -0.1362 | No |
| 14 | Ptpa | 10472 | 0.049 | -0.1517 | No |
| 15 | Hmox2 | 10551 | 0.046 | -0.1509 | No |
| 16 | Msra | 13203 | -0.035 | -0.2601 | No |
| 17 | Gclm | 13294 | -0.038 | -0.2605 | No |
| 18 | Mgst1 | 13461 | -0.045 | -0.2635 | No |
| 19 | Txnrd1 | 13715 | -0.053 | -0.2695 | No |
| 20 | Mpo | 13722 | -0.053 | -0.2650 | No |
| 21 | Sod2 | 14029 | -0.064 | -0.2723 | No |
| 22 | Mbp | 14104 | -0.066 | -0.2694 | No |
| 23 | Prdx6 | 14336 | -0.076 | -0.2724 | No |
| 24 | Ndufs2 | 14674 | -0.087 | -0.2789 | No |
| 25 | Abcc1 | 14869 | -0.094 | -0.2787 | No |
| 26 | Egln2 | 15037 | -0.100 | -0.2768 | No |
| 27 | Lsp1 | 15043 | -0.100 | -0.2680 | No |
| 28 | Junb | 15208 | -0.107 | -0.2653 | No |
| 29 | Sbno2 | 15606 | -0.120 | -0.2714 | No |
| 30 | Glrx2 | 16171 | -0.143 | -0.2824 | No |
| 31 | Hhex | 16184 | -0.143 | -0.2701 | No |
| 32 | Pdlim1 | 17649 | -0.200 | -0.3142 | Yes |
| 33 | Lamtor5 | 17910 | -0.211 | -0.3062 | Yes |
| 34 | Atox1 | 18241 | -0.222 | -0.3002 | Yes |
| 35 | Glrx | 18620 | -0.239 | -0.2947 | Yes |
| 36 | Ercc2 | 18830 | -0.251 | -0.2810 | Yes |
| 37 | Cat | 18922 | -0.256 | -0.2618 | Yes |
| 38 | Gclc | 19653 | -0.291 | -0.2666 | Yes |
| 39 | Ipcef1 | 19786 | -0.299 | -0.2453 | Yes |
| 40 | Cdkn2d | 20640 | -0.345 | -0.2504 | Yes |
| 41 | Txn1 | 20974 | -0.369 | -0.2314 | Yes |
| 42 | Prdx1 | 21135 | -0.381 | -0.2039 | Yes |
| 43 | Selenos | 21508 | -0.410 | -0.1828 | Yes |
| 44 | Sod1 | 22450 | -0.498 | -0.1780 | Yes |
| 45 | Ndufa6 | 22900 | -0.566 | -0.1461 | Yes |
| 46 | Prdx4 | 23153 | -0.624 | -0.1006 | Yes |
| 47 | Prdx2 | 23212 | -0.641 | -0.0454 | Yes |
| 48 | Ftl1 | 23361 | -0.698 | 0.0112 | Yes |