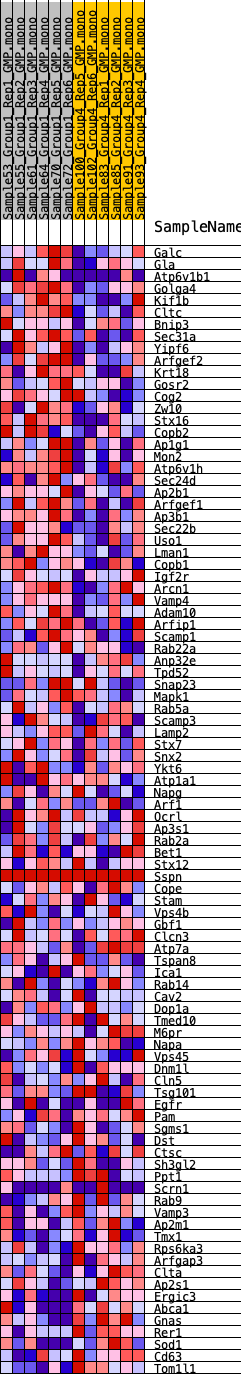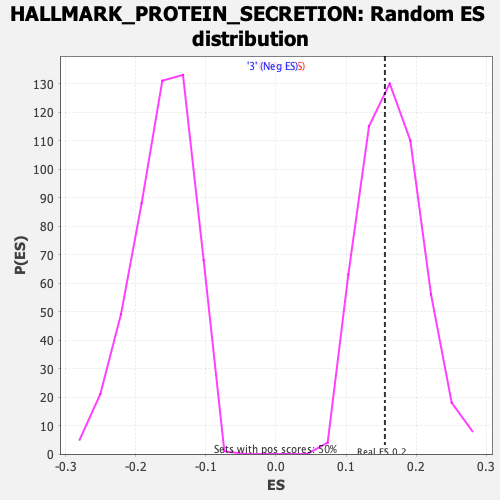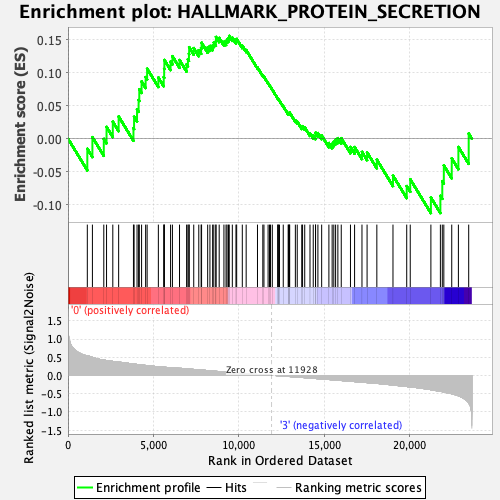
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group4.GMP.mono_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | 0.15553582 |
| Normalized Enrichment Score (NES) | 0.94703704 |
| Nominal p-value | 0.52380955 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Galc | 1130 | 0.537 | -0.0153 | Yes |
| 2 | Gla | 1428 | 0.495 | 0.0022 | Yes |
| 3 | Atp6v1b1 | 2094 | 0.421 | -0.0003 | Yes |
| 4 | Golga4 | 2255 | 0.409 | 0.0178 | Yes |
| 5 | Kif1b | 2621 | 0.384 | 0.0257 | Yes |
| 6 | Cltc | 2963 | 0.367 | 0.0336 | Yes |
| 7 | Bnip3 | 3829 | 0.313 | 0.0159 | Yes |
| 8 | Sec31a | 3866 | 0.311 | 0.0333 | Yes |
| 9 | Yipf6 | 4039 | 0.301 | 0.0444 | Yes |
| 10 | Arfgef2 | 4131 | 0.295 | 0.0585 | Yes |
| 11 | Krt18 | 4171 | 0.294 | 0.0747 | Yes |
| 12 | Gosr2 | 4305 | 0.287 | 0.0866 | Yes |
| 13 | Cog2 | 4542 | 0.274 | 0.0932 | Yes |
| 14 | Zw10 | 4629 | 0.269 | 0.1060 | Yes |
| 15 | Stx16 | 5286 | 0.236 | 0.0925 | Yes |
| 16 | Copb2 | 5601 | 0.228 | 0.0930 | Yes |
| 17 | Ap1g1 | 5626 | 0.226 | 0.1058 | Yes |
| 18 | Mon2 | 5638 | 0.226 | 0.1191 | Yes |
| 19 | Atp6v1h | 6001 | 0.212 | 0.1167 | Yes |
| 20 | Sec24d | 6109 | 0.208 | 0.1248 | Yes |
| 21 | Ap2b1 | 6524 | 0.193 | 0.1190 | Yes |
| 22 | Arfgef1 | 6938 | 0.180 | 0.1124 | Yes |
| 23 | Ap3b1 | 7015 | 0.178 | 0.1200 | Yes |
| 24 | Sec22b | 7075 | 0.175 | 0.1282 | Yes |
| 25 | Uso1 | 7088 | 0.175 | 0.1384 | Yes |
| 26 | Lman1 | 7356 | 0.164 | 0.1371 | Yes |
| 27 | Copb1 | 7651 | 0.154 | 0.1340 | Yes |
| 28 | Igf2r | 7788 | 0.149 | 0.1372 | Yes |
| 29 | Arcn1 | 7815 | 0.147 | 0.1451 | Yes |
| 30 | Vamp4 | 8171 | 0.133 | 0.1381 | Yes |
| 31 | Adam10 | 8303 | 0.128 | 0.1403 | Yes |
| 32 | Arfip1 | 8461 | 0.121 | 0.1411 | Yes |
| 33 | Scamp1 | 8530 | 0.119 | 0.1454 | Yes |
| 34 | Rab22a | 8650 | 0.114 | 0.1473 | Yes |
| 35 | Anp32e | 8657 | 0.113 | 0.1539 | Yes |
| 36 | Tpd52 | 8845 | 0.107 | 0.1525 | Yes |
| 37 | Snap23 | 9122 | 0.097 | 0.1466 | Yes |
| 38 | Mapk1 | 9246 | 0.093 | 0.1471 | Yes |
| 39 | Rab5a | 9306 | 0.091 | 0.1501 | Yes |
| 40 | Scamp3 | 9390 | 0.089 | 0.1520 | Yes |
| 41 | Lamp2 | 9433 | 0.087 | 0.1555 | Yes |
| 42 | Stx7 | 9613 | 0.080 | 0.1528 | No |
| 43 | Snx2 | 9827 | 0.073 | 0.1482 | No |
| 44 | Ykt6 | 9868 | 0.072 | 0.1509 | No |
| 45 | Atp1a1 | 10197 | 0.059 | 0.1406 | No |
| 46 | Napg | 10423 | 0.051 | 0.1341 | No |
| 47 | Arf1 | 11085 | 0.027 | 0.1077 | No |
| 48 | Ocrl | 11397 | 0.018 | 0.0955 | No |
| 49 | Ap3s1 | 11463 | 0.015 | 0.0937 | No |
| 50 | Rab2a | 11706 | 0.008 | 0.0839 | No |
| 51 | Bet1 | 11789 | 0.005 | 0.0807 | No |
| 52 | Stx12 | 11846 | 0.003 | 0.0785 | No |
| 53 | Sspn | 11952 | 0.000 | 0.0740 | No |
| 54 | Cope | 12267 | -0.003 | 0.0608 | No |
| 55 | Stam | 12294 | -0.004 | 0.0599 | No |
| 56 | Vps4b | 12305 | -0.004 | 0.0598 | No |
| 57 | Gbf1 | 12374 | -0.007 | 0.0573 | No |
| 58 | Clcn3 | 12593 | -0.014 | 0.0489 | No |
| 59 | Atp7a | 12880 | -0.024 | 0.0382 | No |
| 60 | Tspan8 | 12902 | -0.025 | 0.0388 | No |
| 61 | Ica1 | 12955 | -0.027 | 0.0383 | No |
| 62 | Rab14 | 12969 | -0.028 | 0.0394 | No |
| 63 | Cav2 | 13301 | -0.039 | 0.0277 | No |
| 64 | Dop1a | 13411 | -0.043 | 0.0257 | No |
| 65 | Tmed10 | 13675 | -0.051 | 0.0176 | No |
| 66 | M6pr | 13723 | -0.053 | 0.0189 | No |
| 67 | Napa | 13849 | -0.057 | 0.0171 | No |
| 68 | Vps45 | 14157 | -0.069 | 0.0082 | No |
| 69 | Dnm1l | 14347 | -0.076 | 0.0048 | No |
| 70 | Cln5 | 14481 | -0.081 | 0.0041 | No |
| 71 | Tsg101 | 14482 | -0.081 | 0.0090 | No |
| 72 | Egfr | 14620 | -0.085 | 0.0084 | No |
| 73 | Pam | 14840 | -0.093 | 0.0047 | No |
| 74 | Sgms1 | 15263 | -0.109 | -0.0066 | No |
| 75 | Dst | 15453 | -0.115 | -0.0076 | No |
| 76 | Ctsc | 15551 | -0.118 | -0.0045 | No |
| 77 | Sh3gl2 | 15655 | -0.123 | -0.0014 | No |
| 78 | Ppt1 | 15788 | -0.128 | 0.0008 | No |
| 79 | Scrn1 | 15986 | -0.136 | 0.0007 | No |
| 80 | Rab9 | 16525 | -0.157 | -0.0126 | No |
| 81 | Vamp3 | 16766 | -0.165 | -0.0127 | No |
| 82 | Ap2m1 | 17198 | -0.181 | -0.0200 | No |
| 83 | Tmx1 | 17497 | -0.194 | -0.0208 | No |
| 84 | Rps6ka3 | 18068 | -0.218 | -0.0318 | No |
| 85 | Arfgap3 | 19012 | -0.261 | -0.0559 | No |
| 86 | Clta | 19816 | -0.300 | -0.0717 | No |
| 87 | Ap2s1 | 20022 | -0.312 | -0.0614 | No |
| 88 | Ergic3 | 21230 | -0.388 | -0.0891 | No |
| 89 | Abca1 | 21785 | -0.429 | -0.0865 | No |
| 90 | Gnas | 21897 | -0.441 | -0.0643 | No |
| 91 | Rer1 | 21987 | -0.452 | -0.0405 | No |
| 92 | Sod1 | 22450 | -0.498 | -0.0298 | No |
| 93 | Cd63 | 22839 | -0.553 | -0.0126 | No |
| 94 | Tom1l1 | 23442 | -0.753 | 0.0077 | No |