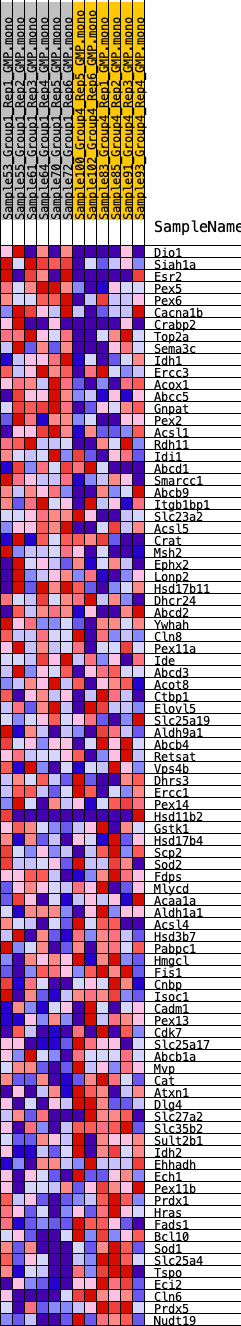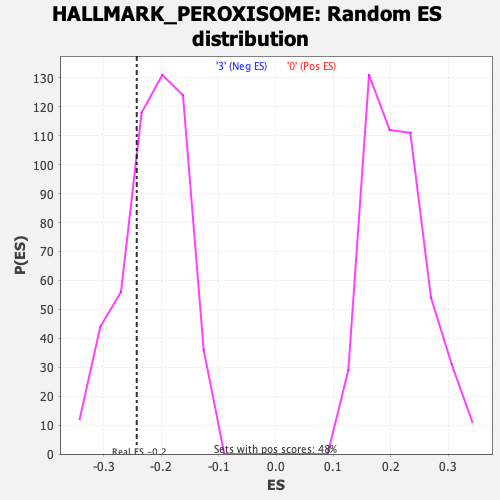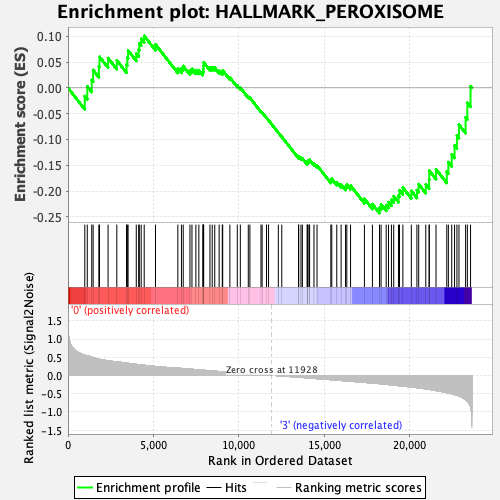
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group4.GMP.mono_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.2427722 |
| Normalized Enrichment Score (NES) | -1.1405139 |
| Nominal p-value | 0.27639157 |
| FDR q-value | 0.48618752 |
| FWER p-Value | 0.954 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dio1 | 982 | 0.551 | -0.0160 | No |
| 2 | Siah1a | 1127 | 0.537 | 0.0030 | No |
| 3 | Esr2 | 1389 | 0.499 | 0.0152 | No |
| 4 | Pex5 | 1470 | 0.488 | 0.0346 | No |
| 5 | Pex6 | 1806 | 0.445 | 0.0412 | No |
| 6 | Cacna1b | 1843 | 0.440 | 0.0602 | No |
| 7 | Crabp2 | 2344 | 0.402 | 0.0577 | No |
| 8 | Top2a | 2856 | 0.370 | 0.0533 | No |
| 9 | Sema3c | 3422 | 0.339 | 0.0451 | No |
| 10 | Idh1 | 3472 | 0.335 | 0.0587 | No |
| 11 | Ercc3 | 3515 | 0.333 | 0.0725 | No |
| 12 | Acox1 | 3994 | 0.304 | 0.0663 | No |
| 13 | Abcc5 | 4135 | 0.295 | 0.0742 | No |
| 14 | Gnpat | 4172 | 0.294 | 0.0864 | No |
| 15 | Pex2 | 4282 | 0.288 | 0.0952 | No |
| 16 | Acsl1 | 4458 | 0.278 | 0.1007 | No |
| 17 | Rdh11 | 5117 | 0.245 | 0.0842 | No |
| 18 | Idi1 | 6426 | 0.197 | 0.0378 | No |
| 19 | Abcd1 | 6636 | 0.193 | 0.0380 | No |
| 20 | Smarcc1 | 6741 | 0.189 | 0.0424 | No |
| 21 | Abcb9 | 7135 | 0.173 | 0.0338 | No |
| 22 | Itgb1bp1 | 7252 | 0.168 | 0.0367 | No |
| 23 | Slc23a2 | 7481 | 0.160 | 0.0345 | No |
| 24 | Acsl5 | 7655 | 0.154 | 0.0343 | No |
| 25 | Crat | 7884 | 0.146 | 0.0314 | No |
| 26 | Msh2 | 7924 | 0.143 | 0.0365 | No |
| 27 | Ephx2 | 7929 | 0.143 | 0.0430 | No |
| 28 | Lonp2 | 7936 | 0.143 | 0.0494 | No |
| 29 | Hsd17b11 | 8304 | 0.128 | 0.0398 | No |
| 30 | Dhcr24 | 8437 | 0.122 | 0.0399 | No |
| 31 | Abcd2 | 8585 | 0.116 | 0.0391 | No |
| 32 | Ywhah | 8842 | 0.107 | 0.0332 | No |
| 33 | Cln8 | 9025 | 0.100 | 0.0301 | No |
| 34 | Pex11a | 9051 | 0.099 | 0.0337 | No |
| 35 | Ide | 9470 | 0.086 | 0.0199 | No |
| 36 | Abcd3 | 9904 | 0.071 | 0.0048 | No |
| 37 | Acot8 | 10082 | 0.063 | 0.0003 | No |
| 38 | Ctbp1 | 10547 | 0.046 | -0.0173 | No |
| 39 | Elovl5 | 10641 | 0.043 | -0.0193 | No |
| 40 | Slc25a19 | 11291 | 0.021 | -0.0458 | No |
| 41 | Aldh9a1 | 11356 | 0.019 | -0.0477 | No |
| 42 | Abcb4 | 11613 | 0.011 | -0.0580 | No |
| 43 | Retsat | 11732 | 0.007 | -0.0628 | No |
| 44 | Vps4b | 12305 | -0.004 | -0.0869 | No |
| 45 | Dhrs3 | 12506 | -0.011 | -0.0948 | No |
| 46 | Ercc1 | 13486 | -0.046 | -0.1343 | No |
| 47 | Pex14 | 13498 | -0.046 | -0.1326 | No |
| 48 | Hsd11b2 | 13630 | -0.051 | -0.1358 | No |
| 49 | Gstk1 | 13712 | -0.052 | -0.1368 | No |
| 50 | Hsd17b4 | 13988 | -0.063 | -0.1456 | No |
| 51 | Scp2 | 14025 | -0.064 | -0.1441 | No |
| 52 | Sod2 | 14029 | -0.064 | -0.1413 | No |
| 53 | Fdps | 14110 | -0.067 | -0.1415 | No |
| 54 | Mlycd | 14135 | -0.068 | -0.1394 | No |
| 55 | Acaa1a | 14390 | -0.078 | -0.1466 | No |
| 56 | Aldh1a1 | 14569 | -0.084 | -0.1502 | No |
| 57 | Acsl4 | 15376 | -0.113 | -0.1792 | No |
| 58 | Hsd3b7 | 15432 | -0.114 | -0.1762 | No |
| 59 | Pabpc1 | 15724 | -0.125 | -0.1827 | No |
| 60 | Hmgcl | 15977 | -0.136 | -0.1871 | No |
| 61 | Fis1 | 16237 | -0.145 | -0.1913 | No |
| 62 | Cnbp | 16312 | -0.149 | -0.1875 | No |
| 63 | Isoc1 | 16530 | -0.157 | -0.1893 | No |
| 64 | Cadm1 | 17341 | -0.187 | -0.2150 | No |
| 65 | Pex13 | 17809 | -0.207 | -0.2252 | No |
| 66 | Cdk7 | 18224 | -0.222 | -0.2324 | Yes |
| 67 | Slc25a17 | 18316 | -0.226 | -0.2257 | Yes |
| 68 | Abcb1a | 18615 | -0.239 | -0.2272 | Yes |
| 69 | Mvp | 18750 | -0.246 | -0.2214 | Yes |
| 70 | Cat | 18922 | -0.256 | -0.2167 | Yes |
| 71 | Atxn1 | 19055 | -0.264 | -0.2100 | Yes |
| 72 | Dlg4 | 19333 | -0.276 | -0.2089 | Yes |
| 73 | Slc27a2 | 19394 | -0.279 | -0.1984 | Yes |
| 74 | Slc35b2 | 19591 | -0.287 | -0.1933 | Yes |
| 75 | Sult2b1 | 20085 | -0.313 | -0.1996 | Yes |
| 76 | Idh2 | 20412 | -0.331 | -0.1980 | Yes |
| 77 | Ehhadh | 20517 | -0.338 | -0.1867 | Yes |
| 78 | Ech1 | 20931 | -0.366 | -0.1871 | Yes |
| 79 | Pex11b | 21126 | -0.380 | -0.1776 | Yes |
| 80 | Prdx1 | 21135 | -0.381 | -0.1602 | Yes |
| 81 | Hras | 21530 | -0.412 | -0.1577 | Yes |
| 82 | Fads1 | 22159 | -0.469 | -0.1625 | Yes |
| 83 | Bcl10 | 22252 | -0.480 | -0.1440 | Yes |
| 84 | Sod1 | 22450 | -0.498 | -0.1291 | Yes |
| 85 | Slc25a4 | 22615 | -0.520 | -0.1118 | Yes |
| 86 | Tspo | 22753 | -0.538 | -0.0924 | Yes |
| 87 | Eci2 | 22872 | -0.559 | -0.0713 | Yes |
| 88 | Cln6 | 23261 | -0.657 | -0.0571 | Yes |
| 89 | Prdx5 | 23363 | -0.699 | -0.0288 | Yes |
| 90 | Nudt19 | 23550 | -0.853 | 0.0031 | Yes |