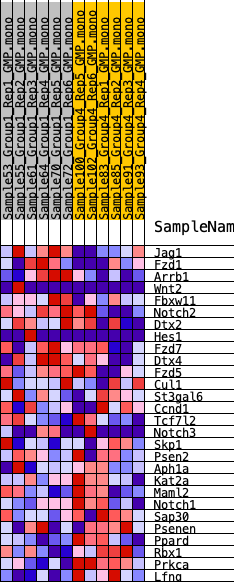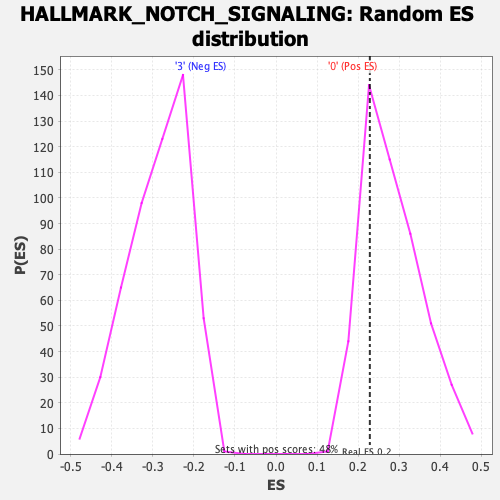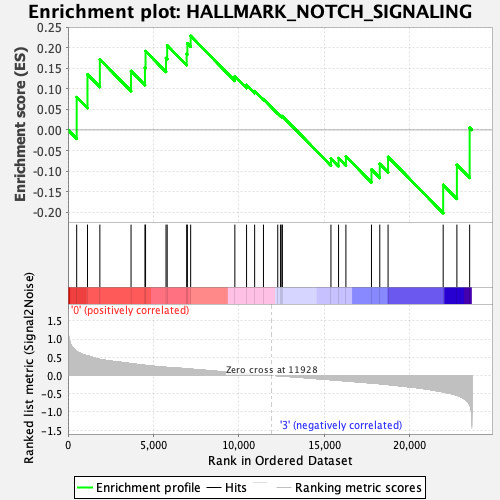
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group4.GMP.mono_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_NOTCH_SIGNALING |
| Enrichment Score (ES) | 0.22861153 |
| Normalized Enrichment Score (NES) | 0.8036066 |
| Nominal p-value | 0.77310926 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Jag1 | 508 | 0.660 | 0.0797 | Yes |
| 2 | Fzd1 | 1140 | 0.534 | 0.1349 | Yes |
| 3 | Arrb1 | 1864 | 0.438 | 0.1715 | Yes |
| 4 | Wnt2 | 3688 | 0.320 | 0.1432 | Yes |
| 5 | Fbxw11 | 4502 | 0.276 | 0.1511 | Yes |
| 6 | Notch2 | 4537 | 0.274 | 0.1916 | Yes |
| 7 | Dtx2 | 5734 | 0.221 | 0.1749 | Yes |
| 8 | Hes1 | 5802 | 0.218 | 0.2054 | Yes |
| 9 | Fzd7 | 6942 | 0.180 | 0.1848 | Yes |
| 10 | Dtx4 | 6984 | 0.178 | 0.2104 | Yes |
| 11 | Fzd5 | 7176 | 0.172 | 0.2286 | Yes |
| 12 | Cul1 | 9761 | 0.075 | 0.1306 | No |
| 13 | St3gal6 | 10446 | 0.050 | 0.1093 | No |
| 14 | Ccnd1 | 10919 | 0.032 | 0.0943 | No |
| 15 | Tcf7l2 | 11435 | 0.017 | 0.0750 | No |
| 16 | Notch3 | 12271 | -0.003 | 0.0400 | No |
| 17 | Skp1 | 12429 | -0.008 | 0.0346 | No |
| 18 | Psen2 | 12513 | -0.011 | 0.0329 | No |
| 19 | Aph1a | 12540 | -0.012 | 0.0336 | No |
| 20 | Kat2a | 15383 | -0.113 | -0.0695 | No |
| 21 | Maml2 | 15828 | -0.130 | -0.0685 | No |
| 22 | Notch1 | 16258 | -0.147 | -0.0641 | No |
| 23 | Sap30 | 17754 | -0.204 | -0.0961 | No |
| 24 | Psenen | 18240 | -0.222 | -0.0826 | No |
| 25 | Ppard | 18725 | -0.244 | -0.0656 | No |
| 26 | Rbx1 | 21949 | -0.447 | -0.1336 | No |
| 27 | Prkca | 22751 | -0.538 | -0.0849 | No |
| 28 | Lfng | 23498 | -0.794 | 0.0053 | No |