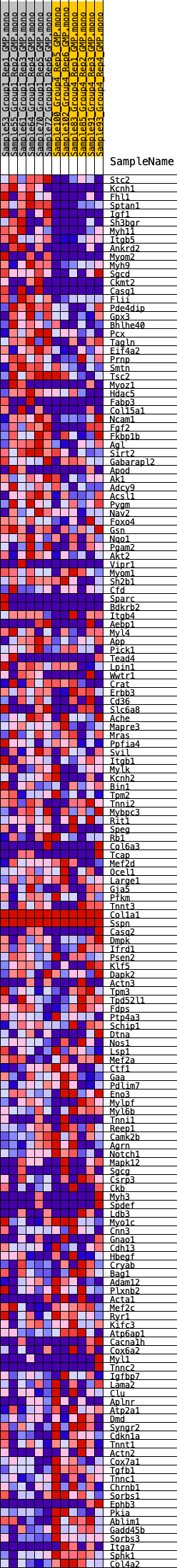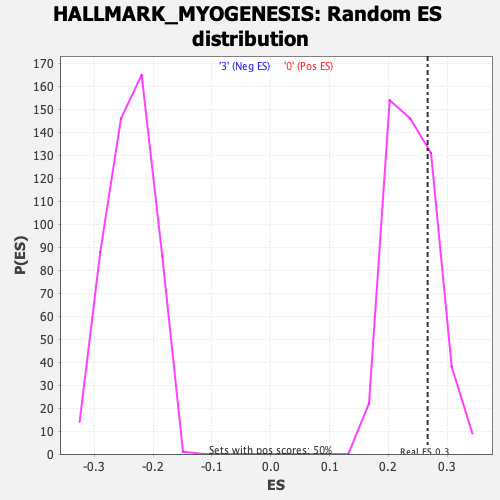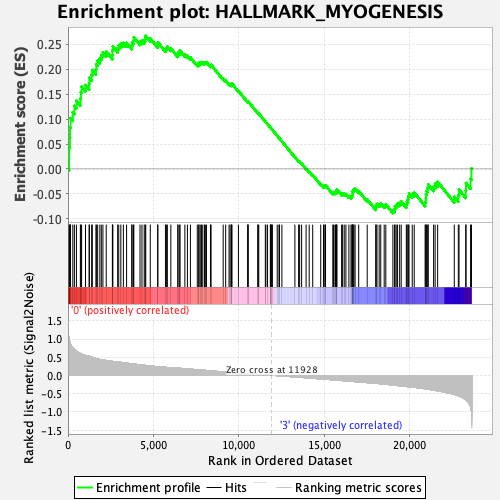
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group4.GMP.mono_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_MYOGENESIS |
| Enrichment Score (ES) | 0.26678237 |
| Normalized Enrichment Score (NES) | 1.111604 |
| Nominal p-value | 0.236 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.966 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Stc2 | 66 | 1.008 | 0.0205 | Yes |
| 2 | Kcnh1 | 68 | 1.003 | 0.0436 | Yes |
| 3 | Fhl1 | 112 | 0.910 | 0.0627 | Yes |
| 4 | Sptan1 | 120 | 0.903 | 0.0833 | Yes |
| 5 | Igf1 | 148 | 0.855 | 0.1019 | Yes |
| 6 | Sh3bgr | 282 | 0.757 | 0.1137 | Yes |
| 7 | Myh11 | 374 | 0.719 | 0.1264 | Yes |
| 8 | Itgb5 | 493 | 0.666 | 0.1368 | Yes |
| 9 | Ankrd2 | 723 | 0.601 | 0.1409 | Yes |
| 10 | Myom2 | 751 | 0.594 | 0.1535 | Yes |
| 11 | Myh9 | 796 | 0.584 | 0.1651 | Yes |
| 12 | Sgcd | 1023 | 0.548 | 0.1681 | Yes |
| 13 | Ckmt2 | 1235 | 0.525 | 0.1712 | Yes |
| 14 | Casq1 | 1251 | 0.523 | 0.1826 | Yes |
| 15 | Flii | 1383 | 0.500 | 0.1886 | Yes |
| 16 | Pde4dip | 1421 | 0.495 | 0.1985 | Yes |
| 17 | Gpx3 | 1638 | 0.464 | 0.2000 | Yes |
| 18 | Bhlhe40 | 1657 | 0.462 | 0.2099 | Yes |
| 19 | Pcx | 1735 | 0.454 | 0.2171 | Yes |
| 20 | Tagln | 1863 | 0.438 | 0.2218 | Yes |
| 21 | Eif4a2 | 1948 | 0.432 | 0.2282 | Yes |
| 22 | Prnp | 2043 | 0.424 | 0.2340 | Yes |
| 23 | Smtn | 2236 | 0.409 | 0.2352 | Yes |
| 24 | Tsc2 | 2596 | 0.386 | 0.2288 | Yes |
| 25 | Myoz1 | 2603 | 0.385 | 0.2375 | Yes |
| 26 | Hdac5 | 2625 | 0.384 | 0.2455 | Yes |
| 27 | Fabp3 | 2906 | 0.369 | 0.2421 | Yes |
| 28 | Col15a1 | 2968 | 0.366 | 0.2479 | Yes |
| 29 | Ncam1 | 3091 | 0.359 | 0.2510 | Yes |
| 30 | Fgf2 | 3239 | 0.350 | 0.2528 | Yes |
| 31 | Fkbp1b | 3428 | 0.338 | 0.2526 | Yes |
| 32 | Agl | 3728 | 0.319 | 0.2472 | Yes |
| 33 | Sirt2 | 3751 | 0.318 | 0.2536 | Yes |
| 34 | Gabarapl2 | 3831 | 0.313 | 0.2575 | Yes |
| 35 | Apod | 3851 | 0.311 | 0.2639 | Yes |
| 36 | Ak1 | 4213 | 0.292 | 0.2552 | Yes |
| 37 | Adcy9 | 4327 | 0.285 | 0.2570 | Yes |
| 38 | Acsl1 | 4458 | 0.278 | 0.2579 | Yes |
| 39 | Pygm | 4518 | 0.275 | 0.2617 | Yes |
| 40 | Nav2 | 4548 | 0.273 | 0.2668 | Yes |
| 41 | Foxo4 | 4813 | 0.260 | 0.2615 | No |
| 42 | Gsn | 5250 | 0.238 | 0.2484 | No |
| 43 | Nqo1 | 5254 | 0.238 | 0.2538 | No |
| 44 | Pgam2 | 5701 | 0.223 | 0.2399 | No |
| 45 | Akt2 | 5756 | 0.220 | 0.2427 | No |
| 46 | Vipr1 | 5812 | 0.218 | 0.2454 | No |
| 47 | Myom1 | 6017 | 0.212 | 0.2416 | No |
| 48 | Sh2b1 | 6422 | 0.197 | 0.2289 | No |
| 49 | Cfd | 6439 | 0.197 | 0.2328 | No |
| 50 | Sparc | 6543 | 0.193 | 0.2328 | No |
| 51 | Bdkrb2 | 6545 | 0.193 | 0.2372 | No |
| 52 | Itgb4 | 6837 | 0.184 | 0.2291 | No |
| 53 | Aebp1 | 6986 | 0.178 | 0.2269 | No |
| 54 | Myl4 | 7162 | 0.172 | 0.2234 | No |
| 55 | App | 7564 | 0.157 | 0.2100 | No |
| 56 | Pick1 | 7650 | 0.154 | 0.2099 | No |
| 57 | Tead4 | 7662 | 0.154 | 0.2130 | No |
| 58 | Lpin1 | 7737 | 0.151 | 0.2133 | No |
| 59 | Wwtr1 | 7814 | 0.147 | 0.2134 | No |
| 60 | Crat | 7884 | 0.146 | 0.2139 | No |
| 61 | Erbb3 | 7983 | 0.141 | 0.2129 | No |
| 62 | Cd36 | 8035 | 0.139 | 0.2140 | No |
| 63 | Slc6a8 | 8100 | 0.135 | 0.2144 | No |
| 64 | Ache | 8336 | 0.126 | 0.2073 | No |
| 65 | Mapre3 | 8370 | 0.125 | 0.2087 | No |
| 66 | Mras | 9078 | 0.099 | 0.1809 | No |
| 67 | Ppfia4 | 9224 | 0.094 | 0.1769 | No |
| 68 | Svil | 9422 | 0.088 | 0.1705 | No |
| 69 | Itgb1 | 9489 | 0.085 | 0.1696 | No |
| 70 | Mylk | 9568 | 0.082 | 0.1682 | No |
| 71 | Kcnh2 | 9574 | 0.082 | 0.1699 | No |
| 72 | Bin1 | 9587 | 0.081 | 0.1712 | No |
| 73 | Tpm2 | 9970 | 0.067 | 0.1565 | No |
| 74 | Tnni2 | 10514 | 0.047 | 0.1345 | No |
| 75 | Mybpc3 | 10532 | 0.047 | 0.1348 | No |
| 76 | Rit1 | 10535 | 0.047 | 0.1358 | No |
| 77 | Speg | 11102 | 0.026 | 0.1123 | No |
| 78 | Rb1 | 11120 | 0.026 | 0.1122 | No |
| 79 | Col6a3 | 11160 | 0.025 | 0.1111 | No |
| 80 | Tcap | 11544 | 0.013 | 0.0950 | No |
| 81 | Mef2d | 11577 | 0.012 | 0.0940 | No |
| 82 | Ocel1 | 11674 | 0.008 | 0.0901 | No |
| 83 | Large1 | 11838 | 0.003 | 0.0832 | No |
| 84 | Gja5 | 11851 | 0.002 | 0.0827 | No |
| 85 | Pfkm | 11856 | 0.002 | 0.0826 | No |
| 86 | Tnnt3 | 11917 | 0.000 | 0.0801 | No |
| 87 | Col1a1 | 11928 | 0.000 | 0.0796 | No |
| 88 | Sspn | 11952 | 0.000 | 0.0786 | No |
| 89 | Casq2 | 12240 | -0.002 | 0.0665 | No |
| 90 | Dmpk | 12342 | -0.006 | 0.0623 | No |
| 91 | Ifrd1 | 12373 | -0.007 | 0.0612 | No |
| 92 | Psen2 | 12513 | -0.011 | 0.0555 | No |
| 93 | Klf5 | 13274 | -0.038 | 0.0240 | No |
| 94 | Dapk2 | 13491 | -0.046 | 0.0158 | No |
| 95 | Actn3 | 13550 | -0.048 | 0.0145 | No |
| 96 | Tpm3 | 13667 | -0.051 | 0.0107 | No |
| 97 | Tpd52l1 | 13929 | -0.060 | 0.0010 | No |
| 98 | Fdps | 14110 | -0.067 | -0.0052 | No |
| 99 | Ptp4a3 | 14316 | -0.075 | -0.0122 | No |
| 100 | Schip1 | 14785 | -0.091 | -0.0300 | No |
| 101 | Dtna | 14946 | -0.096 | -0.0346 | No |
| 102 | Nos1 | 14981 | -0.098 | -0.0338 | No |
| 103 | Lsp1 | 15043 | -0.100 | -0.0341 | No |
| 104 | Mef2a | 15066 | -0.101 | -0.0327 | No |
| 105 | Ctf1 | 15493 | -0.116 | -0.0482 | No |
| 106 | Gaa | 15528 | -0.118 | -0.0469 | No |
| 107 | Pdlim7 | 15615 | -0.121 | -0.0478 | No |
| 108 | Eno3 | 15651 | -0.122 | -0.0465 | No |
| 109 | Mylpf | 15714 | -0.125 | -0.0462 | No |
| 110 | Myl6b | 15716 | -0.125 | -0.0434 | No |
| 111 | Tnni1 | 15728 | -0.125 | -0.0409 | No |
| 112 | Reep1 | 16007 | -0.137 | -0.0496 | No |
| 113 | Camk2b | 16066 | -0.139 | -0.0489 | No |
| 114 | Agrn | 16160 | -0.142 | -0.0496 | No |
| 115 | Notch1 | 16258 | -0.147 | -0.0503 | No |
| 116 | Mapk12 | 16433 | -0.154 | -0.0542 | No |
| 117 | Sgcg | 16556 | -0.158 | -0.0557 | No |
| 118 | Csrp3 | 16609 | -0.160 | -0.0543 | No |
| 119 | Ckb | 16638 | -0.161 | -0.0517 | No |
| 120 | Myh3 | 16647 | -0.162 | -0.0483 | No |
| 121 | Spdef | 16648 | -0.162 | -0.0446 | No |
| 122 | Ldb3 | 16691 | -0.162 | -0.0426 | No |
| 123 | Myo1c | 16728 | -0.164 | -0.0404 | No |
| 124 | Cnn3 | 16796 | -0.166 | -0.0394 | No |
| 125 | Gnao1 | 17006 | -0.175 | -0.0443 | No |
| 126 | Cdh13 | 17504 | -0.194 | -0.0610 | No |
| 127 | Hbegf | 18004 | -0.215 | -0.0773 | No |
| 128 | Cryab | 18023 | -0.217 | -0.0731 | No |
| 129 | Bag1 | 18076 | -0.218 | -0.0702 | No |
| 130 | Adam12 | 18205 | -0.221 | -0.0706 | No |
| 131 | Plxnb2 | 18297 | -0.225 | -0.0693 | No |
| 132 | Acta1 | 18498 | -0.234 | -0.0724 | No |
| 133 | Mef2c | 18593 | -0.238 | -0.0709 | No |
| 134 | Ryr1 | 19008 | -0.261 | -0.0826 | No |
| 135 | Kifc3 | 19110 | -0.266 | -0.0807 | No |
| 136 | Atp6ap1 | 19127 | -0.267 | -0.0752 | No |
| 137 | Cacna1h | 19200 | -0.270 | -0.0721 | No |
| 138 | Cox6a2 | 19280 | -0.273 | -0.0691 | No |
| 139 | Myl1 | 19403 | -0.280 | -0.0679 | No |
| 140 | Tnnc2 | 19494 | -0.285 | -0.0651 | No |
| 141 | Igfbp7 | 19791 | -0.300 | -0.0708 | No |
| 142 | Lama2 | 19829 | -0.301 | -0.0654 | No |
| 143 | Clu | 19888 | -0.306 | -0.0609 | No |
| 144 | Aplnr | 19907 | -0.306 | -0.0546 | No |
| 145 | Atp2a1 | 19951 | -0.309 | -0.0493 | No |
| 146 | Dmd | 20139 | -0.316 | -0.0499 | No |
| 147 | Syngr2 | 20255 | -0.323 | -0.0474 | No |
| 148 | Cdkn1a | 20892 | -0.363 | -0.0661 | No |
| 149 | Tnnt1 | 20938 | -0.367 | -0.0596 | No |
| 150 | Actn2 | 20945 | -0.367 | -0.0514 | No |
| 151 | Cox7a1 | 20982 | -0.369 | -0.0444 | No |
| 152 | Tgfb1 | 21043 | -0.373 | -0.0383 | No |
| 153 | Tnnc1 | 21076 | -0.376 | -0.0310 | No |
| 154 | Chrnb1 | 21389 | -0.400 | -0.0351 | No |
| 155 | Sorbs1 | 21478 | -0.407 | -0.0294 | No |
| 156 | Ephb3 | 21625 | -0.420 | -0.0259 | No |
| 157 | Pkia | 22598 | -0.518 | -0.0554 | No |
| 158 | Ablim1 | 22834 | -0.552 | -0.0527 | No |
| 159 | Gadd45b | 22876 | -0.561 | -0.0415 | No |
| 160 | Sorbs3 | 23270 | -0.662 | -0.0429 | No |
| 161 | Itga7 | 23285 | -0.667 | -0.0281 | No |
| 162 | Sphk1 | 23552 | -0.854 | -0.0198 | No |
| 163 | Col4a2 | 23604 | -0.987 | 0.0009 | No |