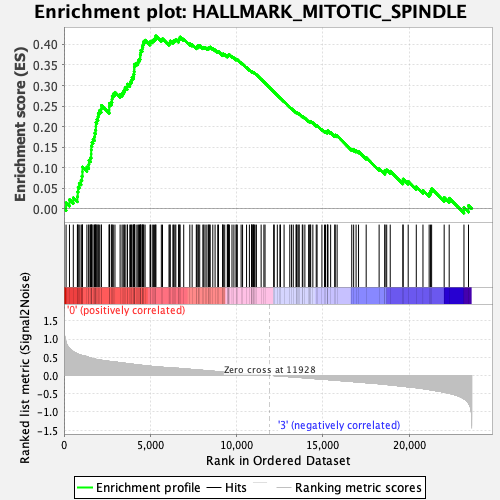
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group4.GMP.mono_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_MITOTIC_SPINDLE |
| Enrichment Score (ES) | 0.42097718 |
| Normalized Enrichment Score (NES) | 1.882951 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.014362385 |
| FWER p-Value | 0.02 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Sptan1 | 120 | 0.903 | 0.0144 | Yes |
| 2 | Sun2 | 317 | 0.740 | 0.0220 | Yes |
| 3 | Cenpf | 539 | 0.648 | 0.0266 | Yes |
| 4 | Incenp | 776 | 0.588 | 0.0292 | Yes |
| 5 | Myh9 | 796 | 0.584 | 0.0410 | Yes |
| 6 | Arhgef12 | 818 | 0.577 | 0.0526 | Yes |
| 7 | Espl1 | 881 | 0.568 | 0.0622 | Yes |
| 8 | Dlgap5 | 989 | 0.550 | 0.0695 | Yes |
| 9 | Racgap1 | 1039 | 0.547 | 0.0792 | Yes |
| 10 | Cntrob | 1068 | 0.543 | 0.0898 | Yes |
| 11 | Anln | 1070 | 0.543 | 0.1015 | Yes |
| 12 | Kif2c | 1327 | 0.509 | 0.1015 | Yes |
| 13 | Kif11 | 1435 | 0.494 | 0.1076 | Yes |
| 14 | Ckap5 | 1446 | 0.492 | 0.1178 | Yes |
| 15 | Cdk5rap2 | 1540 | 0.478 | 0.1242 | Yes |
| 16 | Kif23 | 1572 | 0.473 | 0.1331 | Yes |
| 17 | Cep72 | 1575 | 0.473 | 0.1432 | Yes |
| 18 | Sptbn1 | 1583 | 0.472 | 0.1531 | Yes |
| 19 | Hdac6 | 1620 | 0.466 | 0.1616 | Yes |
| 20 | Cep250 | 1684 | 0.460 | 0.1689 | Yes |
| 21 | Prc1 | 1766 | 0.449 | 0.1751 | Yes |
| 22 | Kntc1 | 1784 | 0.447 | 0.1841 | Yes |
| 23 | Cdc42ep4 | 1833 | 0.442 | 0.1916 | Yes |
| 24 | Nin | 1847 | 0.440 | 0.2005 | Yes |
| 25 | Dynll2 | 1850 | 0.440 | 0.2099 | Yes |
| 26 | Ect2 | 1906 | 0.435 | 0.2170 | Yes |
| 27 | Trio | 1969 | 0.430 | 0.2236 | Yes |
| 28 | Shroom1 | 1983 | 0.429 | 0.2323 | Yes |
| 29 | Pcnt | 2047 | 0.424 | 0.2388 | Yes |
| 30 | Numa1 | 2156 | 0.415 | 0.2432 | Yes |
| 31 | Flna | 2167 | 0.414 | 0.2517 | Yes |
| 32 | Kif1b | 2621 | 0.384 | 0.2406 | Yes |
| 33 | Kif22 | 2622 | 0.384 | 0.2489 | Yes |
| 34 | Nek2 | 2633 | 0.383 | 0.2568 | Yes |
| 35 | Ttk | 2742 | 0.379 | 0.2603 | Yes |
| 36 | Cenpe | 2782 | 0.375 | 0.2668 | Yes |
| 37 | Tubgcp3 | 2791 | 0.374 | 0.2745 | Yes |
| 38 | Top2a | 2856 | 0.370 | 0.2798 | Yes |
| 39 | Tubgcp6 | 2953 | 0.368 | 0.2836 | Yes |
| 40 | Ophn1 | 3252 | 0.349 | 0.2784 | Yes |
| 41 | Stk38l | 3364 | 0.342 | 0.2811 | Yes |
| 42 | Cep131 | 3433 | 0.338 | 0.2855 | Yes |
| 43 | Lrpprc | 3497 | 0.333 | 0.2900 | Yes |
| 44 | Dync1h1 | 3543 | 0.331 | 0.2952 | Yes |
| 45 | Cdk1 | 3665 | 0.321 | 0.2970 | Yes |
| 46 | Als2 | 3675 | 0.320 | 0.3035 | Yes |
| 47 | Gemin4 | 3815 | 0.314 | 0.3044 | Yes |
| 48 | Ccnb2 | 3855 | 0.311 | 0.3095 | Yes |
| 49 | Cntrl | 3919 | 0.308 | 0.3134 | Yes |
| 50 | Pdlim5 | 3940 | 0.306 | 0.3192 | Yes |
| 51 | Kif4 | 4021 | 0.302 | 0.3223 | Yes |
| 52 | Cep192 | 4045 | 0.301 | 0.3278 | Yes |
| 53 | Ndc80 | 4062 | 0.300 | 0.3336 | Yes |
| 54 | Smc1a | 4072 | 0.299 | 0.3397 | Yes |
| 55 | Lmnb1 | 4073 | 0.299 | 0.3461 | Yes |
| 56 | Bub1 | 4087 | 0.298 | 0.3520 | Yes |
| 57 | Epb41l2 | 4180 | 0.293 | 0.3544 | Yes |
| 58 | Aurka | 4280 | 0.288 | 0.3564 | Yes |
| 59 | Arhgap4 | 4309 | 0.286 | 0.3614 | Yes |
| 60 | Kif20b | 4375 | 0.282 | 0.3647 | Yes |
| 61 | Plk1 | 4416 | 0.280 | 0.3691 | Yes |
| 62 | Ranbp9 | 4417 | 0.280 | 0.3751 | Yes |
| 63 | Smc3 | 4444 | 0.279 | 0.3800 | Yes |
| 64 | Arhgef7 | 4449 | 0.279 | 0.3859 | Yes |
| 65 | Rasa1 | 4532 | 0.274 | 0.3883 | Yes |
| 66 | Notch2 | 4537 | 0.274 | 0.3941 | Yes |
| 67 | Fscn1 | 4570 | 0.272 | 0.3986 | Yes |
| 68 | Vcl | 4594 | 0.271 | 0.4034 | Yes |
| 69 | Arap3 | 4626 | 0.269 | 0.4079 | Yes |
| 70 | Arhgef2 | 4699 | 0.265 | 0.4106 | Yes |
| 71 | Stau1 | 4987 | 0.251 | 0.4038 | Yes |
| 72 | Rasa2 | 5015 | 0.250 | 0.4080 | Yes |
| 73 | Klc1 | 5113 | 0.245 | 0.4092 | Yes |
| 74 | Cenpj | 5182 | 0.241 | 0.4115 | Yes |
| 75 | Gsn | 5250 | 0.238 | 0.4138 | Yes |
| 76 | Kif15 | 5314 | 0.235 | 0.4162 | Yes |
| 77 | Tsc1 | 5321 | 0.235 | 0.4210 | Yes |
| 78 | Tpx2 | 5641 | 0.226 | 0.4122 | No |
| 79 | Sorbs2 | 5709 | 0.222 | 0.4142 | No |
| 80 | Fgd4 | 6086 | 0.209 | 0.4026 | No |
| 81 | Cyth2 | 6135 | 0.206 | 0.4051 | No |
| 82 | Arhgap10 | 6155 | 0.206 | 0.4087 | No |
| 83 | Septin9 | 6311 | 0.202 | 0.4064 | No |
| 84 | Bcr | 6354 | 0.200 | 0.4090 | No |
| 85 | Hook3 | 6423 | 0.197 | 0.4103 | No |
| 86 | Myh10 | 6480 | 0.195 | 0.4121 | No |
| 87 | Tubgcp5 | 6642 | 0.193 | 0.4094 | No |
| 88 | Alms1 | 6657 | 0.192 | 0.4130 | No |
| 89 | Prex1 | 6704 | 0.190 | 0.4151 | No |
| 90 | Cdc42bpa | 6727 | 0.190 | 0.4183 | No |
| 91 | Arfgef1 | 6938 | 0.180 | 0.4132 | No |
| 92 | Farp1 | 7292 | 0.167 | 0.4017 | No |
| 93 | Tiam1 | 7426 | 0.161 | 0.3996 | No |
| 94 | Nedd9 | 7667 | 0.154 | 0.3926 | No |
| 95 | Mid1 | 7698 | 0.152 | 0.3946 | No |
| 96 | Wasf1 | 7772 | 0.149 | 0.3947 | No |
| 97 | Tlk1 | 7777 | 0.149 | 0.3978 | No |
| 98 | Sass6 | 7865 | 0.146 | 0.3972 | No |
| 99 | Myo9b | 8048 | 0.138 | 0.3924 | No |
| 100 | Nf1 | 8099 | 0.135 | 0.3932 | No |
| 101 | Sos1 | 8176 | 0.132 | 0.3928 | No |
| 102 | Dock4 | 8280 | 0.128 | 0.3912 | No |
| 103 | Nck2 | 8367 | 0.125 | 0.3902 | No |
| 104 | Pafah1b1 | 8392 | 0.124 | 0.3919 | No |
| 105 | Rfc1 | 8441 | 0.122 | 0.3925 | No |
| 106 | Flnb | 8471 | 0.121 | 0.3939 | No |
| 107 | Map3k11 | 8624 | 0.115 | 0.3899 | No |
| 108 | Dock2 | 8756 | 0.109 | 0.3866 | No |
| 109 | Arfip2 | 8910 | 0.105 | 0.3824 | No |
| 110 | Dlg1 | 8954 | 0.103 | 0.3828 | No |
| 111 | Mark4 | 9195 | 0.095 | 0.3746 | No |
| 112 | Actn4 | 9202 | 0.094 | 0.3763 | No |
| 113 | Wasl | 9227 | 0.094 | 0.3773 | No |
| 114 | Clip1 | 9315 | 0.091 | 0.3756 | No |
| 115 | Lats1 | 9463 | 0.086 | 0.3712 | No |
| 116 | Pkd2 | 9481 | 0.085 | 0.3723 | No |
| 117 | Taok2 | 9518 | 0.084 | 0.3726 | No |
| 118 | Arhgef11 | 9523 | 0.083 | 0.3742 | No |
| 119 | Net1 | 9544 | 0.083 | 0.3751 | No |
| 120 | Bin1 | 9587 | 0.081 | 0.3751 | No |
| 121 | Abi1 | 9736 | 0.076 | 0.3704 | No |
| 122 | Rictor | 9847 | 0.073 | 0.3673 | No |
| 123 | Rapgef6 | 9971 | 0.067 | 0.3635 | No |
| 124 | Pcm1 | 9988 | 0.067 | 0.3642 | No |
| 125 | Arhgap29 | 10045 | 0.064 | 0.3632 | No |
| 126 | Brca2 | 10265 | 0.057 | 0.3551 | No |
| 127 | Arhgef3 | 10349 | 0.054 | 0.3527 | No |
| 128 | Nusap1 | 10580 | 0.045 | 0.3439 | No |
| 129 | Ppp4r2 | 10745 | 0.039 | 0.3377 | No |
| 130 | Abl1 | 10871 | 0.034 | 0.3331 | No |
| 131 | Nck1 | 10877 | 0.034 | 0.3336 | No |
| 132 | Ywhae | 10885 | 0.033 | 0.3341 | No |
| 133 | Clasp1 | 10941 | 0.031 | 0.3324 | No |
| 134 | Rab3gap1 | 10974 | 0.030 | 0.3317 | No |
| 135 | Ccdc88a | 10976 | 0.030 | 0.3323 | No |
| 136 | Pif1 | 11031 | 0.029 | 0.3306 | No |
| 137 | Cdc27 | 11071 | 0.027 | 0.3295 | No |
| 138 | Rhot2 | 11150 | 0.025 | 0.3267 | No |
| 139 | Plekhg2 | 11425 | 0.017 | 0.3154 | No |
| 140 | Smc4 | 11585 | 0.012 | 0.3089 | No |
| 141 | Capzb | 11651 | 0.009 | 0.3063 | No |
| 142 | Kptn | 12152 | -0.000 | 0.2850 | No |
| 143 | Arhgap5 | 12184 | -0.001 | 0.2837 | No |
| 144 | Kif3b | 12363 | -0.006 | 0.2762 | No |
| 145 | Mid1ip1 | 12514 | -0.011 | 0.2700 | No |
| 146 | Katna1 | 12546 | -0.012 | 0.2690 | No |
| 147 | Kif5b | 12755 | -0.019 | 0.2605 | No |
| 148 | Rock1 | 13086 | -0.030 | 0.2471 | No |
| 149 | Tbcd | 13188 | -0.035 | 0.2435 | No |
| 150 | Cttn | 13287 | -0.038 | 0.2402 | No |
| 151 | Itsn1 | 13449 | -0.044 | 0.2343 | No |
| 152 | Akap13 | 13471 | -0.045 | 0.2343 | No |
| 153 | Myo1e | 13512 | -0.046 | 0.2336 | No |
| 154 | Katnb1 | 13610 | -0.050 | 0.2306 | No |
| 155 | Abr | 13626 | -0.051 | 0.2310 | No |
| 156 | Ezr | 13811 | -0.057 | 0.2244 | No |
| 157 | Rabgap1 | 13843 | -0.057 | 0.2243 | No |
| 158 | Cep57 | 13960 | -0.061 | 0.2207 | No |
| 159 | Tubgcp2 | 14177 | -0.070 | 0.2130 | No |
| 160 | Mapre1 | 14248 | -0.072 | 0.2115 | No |
| 161 | Kifap3 | 14256 | -0.072 | 0.2128 | No |
| 162 | Arf6 | 14297 | -0.074 | 0.2127 | No |
| 163 | Pcgf5 | 14423 | -0.079 | 0.2091 | No |
| 164 | Palld | 14618 | -0.085 | 0.2026 | No |
| 165 | Apc | 14667 | -0.087 | 0.2024 | No |
| 166 | Csnk1d | 14948 | -0.096 | 0.1926 | No |
| 167 | Wasf2 | 15099 | -0.102 | 0.1884 | No |
| 168 | Fbxo5 | 15155 | -0.105 | 0.1883 | No |
| 169 | Cdc42 | 15262 | -0.109 | 0.1861 | No |
| 170 | Rasal2 | 15286 | -0.110 | 0.1875 | No |
| 171 | Map1s | 15288 | -0.110 | 0.1898 | No |
| 172 | Dst | 15453 | -0.115 | 0.1853 | No |
| 173 | Epb41 | 15691 | -0.124 | 0.1779 | No |
| 174 | Clip2 | 15718 | -0.125 | 0.1795 | No |
| 175 | Shroom2 | 15829 | -0.130 | 0.1776 | No |
| 176 | Atg4b | 16677 | -0.162 | 0.1449 | No |
| 177 | Ssh2 | 16787 | -0.166 | 0.1438 | No |
| 178 | Pxn | 16928 | -0.171 | 0.1416 | No |
| 179 | Cdc42ep2 | 17070 | -0.177 | 0.1394 | No |
| 180 | Synpo | 17513 | -0.194 | 0.1247 | No |
| 181 | Birc5 | 18263 | -0.223 | 0.0976 | No |
| 182 | Arhgap27 | 18596 | -0.238 | 0.0885 | No |
| 183 | Arl8a | 18600 | -0.239 | 0.0936 | No |
| 184 | Ralbp1 | 18702 | -0.244 | 0.0945 | No |
| 185 | Tubd1 | 18907 | -0.255 | 0.0913 | No |
| 186 | Tuba4a | 19643 | -0.291 | 0.0662 | No |
| 187 | Rhof | 19652 | -0.291 | 0.0722 | No |
| 188 | Cd2ap | 19948 | -0.308 | 0.0662 | No |
| 189 | Arhgdia | 20417 | -0.332 | 0.0534 | No |
| 190 | Kif3c | 20804 | -0.356 | 0.0447 | No |
| 191 | Rapgef5 | 21163 | -0.382 | 0.0376 | No |
| 192 | Marcks | 21244 | -0.389 | 0.0426 | No |
| 193 | Fgd6 | 21306 | -0.393 | 0.0485 | No |
| 194 | Bcl2l11 | 22033 | -0.455 | 0.0273 | No |
| 195 | Sac3d1 | 22324 | -0.486 | 0.0255 | No |
| 196 | Uxt | 23180 | -0.633 | 0.0026 | No |
| 197 | Llgl1 | 23443 | -0.754 | 0.0077 | No |